Multicenter Study of Cronobacter sakazakii Infections in Humans, Europe, 2017
- PMID: 30789137
- PMCID: PMC6390735
- DOI: 10.3201/eid2503.181652
Multicenter Study of Cronobacter sakazakii Infections in Humans, Europe, 2017
Abstract
Cronobacter sakazakii has been documented as a cause of life-threating infections, predominantly in neonates. We conducted a multicenter study to assess the occurrence of C. sakazakii across Europe and the extent of clonality for outbreak detection. National coordinators representing 24 countries in Europe were requested to submit all human C. sakazakii isolates collected during 2017 to a study center in Austria. Testing at the center included species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, subtyping by whole-genome sequencing (WGS), and determination of antimicrobial resistance. Eleven countries sent 77 isolates, including 36 isolates from 2017 and 41 historical isolates. Fifty-nine isolates were confirmed as C. sakazakii by WGS, highlighting the challenge of correctly identifying Cronobacter spp. WGS-based typing revealed high strain diversity, indicating absence of multinational outbreaks in 2017, but identified 4 previously unpublished historical outbreaks. WGS is the recommended method for accurate identification, typing, and detection of this pathogen.
Keywords: Cronobacter sakazakii; Europe; bacteria; bacterial typing; food safety; foodborne diseases; outbreak investigation; public health surveillance; whole-genome sequencing.
Figures

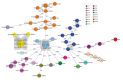
References
-
- Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, et al. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol. 2008;58:1442–7. 10.1099/ijs.0.65577-0 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

