A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases
- PMID: 30789718
- PMCID: PMC6503667
- DOI: 10.1021/acs.biochem.9b00044
A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases
Abstract
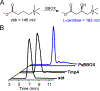
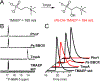
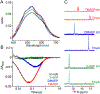
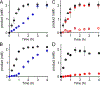
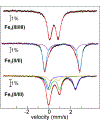
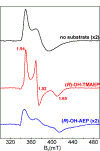
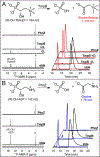
The assignment of biochemical functions to hypothetical proteins is challenged by functional diversification within many protein structural superfamilies. This diversification, which is particularly common for metalloenzymes, renders functional annotations that are founded solely on sequence and domain similarities unreliable and often erroneous. Definitive biochemical characterization to delineate functional subgroups within these superfamilies will aid in improving bioinformatic approaches for functional annotation. We describe here the structural and functional characterization of two non-heme-iron oxygenases, TmpA and TmpB, which are encoded by a genomically clustered pair of genes found in more than 350 species of bacteria. TmpA and TmpB are functional homologues of a pair of enzymes (PhnY and PhnZ) that degrade 2-aminoethylphosphonate but instead act on its naturally occurring, quaternary ammonium analogue, 2-(trimethylammonio)ethylphosphonate (TMAEP). TmpA, an iron(II)- and 2-(oxo)glutarate-dependent oxygenase misannotated as a γ-butyrobetaine (γbb) hydroxylase, shows no activity toward γbb but efficiently hydroxylates TMAEP. The product, ( R)-1-hydroxy-2-(trimethylammonio)ethylphosphonate [( R)-OH-TMAEP], then serves as the substrate for the second enzyme, TmpB. By contrast to its purported phosphohydrolytic activity, TmpB is an HD-domain oxygenase that uses a mixed-valent diiron cofactor to enact oxidative cleavage of the C-P bond of its substrate, yielding glycine betaine and phosphate. The high specificities of TmpA and TmpB for their N-trimethylated substrates suggest that they have evolved specifically to degrade TMAEP, which was not previously known to be subject to microbial catabolism. This study thus adds to the growing list of known pathways through which microbes break down organophosphonates to harvest phosphorus, carbon, and nitrogen in nutrient-limited niches.
Figures

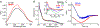

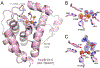
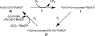
References
-
- Sono M, Roach MP, Coulter ED, Dawson JH (1996) Heme-containing oxygenases. Chem. Rev 96, 2841–2888. - PubMed
-
- Hausinger RP (2015) 2-Oxoglutarate-Dependent Oxygenases, Hausinger RP, Schofield CJ, Ed., The Royal Society of Chemistry.
-
- Andrews SC (2010) The ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta 1800, 691–705. - PubMed
-
- Aravind L, Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci 23, 469–472. - PubMed
-
- Baier F, Copp JN, Tokuriki N (2016) Evolution of enzyme superfamilies: Comprehensive exploration of sequence-function relationships. Biochemistry 55, 6375–6388. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

