Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development
- PMID: 30789893
- PMCID: PMC6400406
- DOI: 10.1371/journal.pbio.3000152
Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development
Abstract
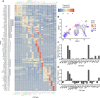
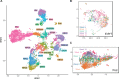
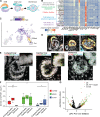
The current understanding of mammalian kidney development is largely based on mouse models. Recent landmark studies revealed pervasive differences in renal embryogenesis between mouse and human. The scarcity of detailed gene expression data in humans therefore hampers a thorough understanding of human kidney development and the possible developmental origin of kidney diseases. In this paper, we present a single-cell transcriptomics study of the human fetal kidney. We identified 22 cell types and a host of marker genes. Comparison of samples from different developmental ages revealed continuous gene expression changes in podocytes. To demonstrate the usefulness of our data set, we explored the heterogeneity of the nephrogenic niche, localized podocyte precursors, and confirmed disease-associated marker genes. With close to 18,000 renal cells from five different developmental ages, this study provides a rich resource for the elucidation of human kidney development, easily accessible through an interactive web application.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

