The Antiresection Activity of the X Protein Encoded by Hepatitis Virus B
- PMID: 30791110
- PMCID: PMC6618260
- DOI: 10.1002/hep.30571
The Antiresection Activity of the X Protein Encoded by Hepatitis Virus B
Abstract
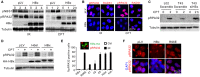
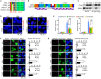
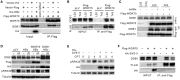
Chronic infection of hepatitis B virus (HBV) is associated with an increased incidence of hepatocellular carcinoma (HCC). HBV encodes an oncoprotein, hepatitis B x protein (HBx), that is crucial for viral replication and interferes with multiple cellular activities including gene expression, histone modifications, and genomic stability. To date, it remains unclear how disruption of these activities contributes to hepatocarcinogenesis. Here, we report that HBV exhibits antiresection activity by disrupting DNA end resection, thus impairing the initial steps of homologous recombination (HR). This antiresection activity occurs in primary human hepatocytes undergoing a natural viral infection-replication cycle as well as in cells with integrated HBV genomes. Among the seven HBV-encoded proteins, we identified HBx as the sole viral factor that inhibits resection. By disrupting an evolutionarily conserved Cullin4A-damage-specific DNA binding protein 1-RING type of E3 ligase, CRL4WDR70 , through its H-box, we show that HBx inhibits H2B monoubiquitylation at lysine 120 at double-strand breaks, thus reducing the efficiency of long-range resection. We further show that directly impairing H2B monoubiquitylation elicited tumorigenesis upon engraftment of deficient cells in athymic mice, confirming that the impairment of CRL4WDR70 function by HBx is sufficient to promote carcinogenesis. Finally, we demonstrate that lack of H2B monoubiquitylation is manifest in human HBV-associated HCC when compared with HBV-free HCC, implying corresponding defects of epigenetic regulation and end resection. Conclusion: The antiresection activity of HBx induces an HR defect and genomic instability and contributes to tumorigenesis of host hepatocytes.
© 2019 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of American Association for the Study of Liver Diseases.
Figures





References
-
- El‐Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118‐1127. - PubMed
-
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674‐687. - PubMed
-
- Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22,707 men in Taiwan. Lancet 1981;2:1129‐1133. - PubMed
-
- Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature 1980;286:533‐535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2013CB911000/Ministry of Science and Technology of the People's Republic of China/International
- 201703D321013/Department of Science and Technology of Shanxi Province/International
- 2017FZ0034/Department of Science and Technology of Sichuan Province/International
- 31471276/National Natural Science Foundation of China/International
- 31771580/National Natural Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

