Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)₂dppz]2+ and [Ru(phen)₂dppz]2+ with a DNA Molecule
- PMID: 30791625
- PMCID: PMC6412511
- DOI: 10.3390/molecules24040769
Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)₂dppz]2+ and [Ru(phen)₂dppz]2+ with a DNA Molecule
Abstract
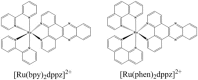
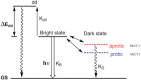
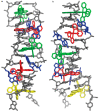
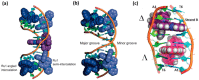
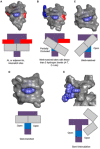
[Ru(bpy)₂dppz]2+ and [Ru(phen)₂dppz]2+ as the light switches of the deoxyribose nucleic acid (DNA) molecule have attracted much attention and have become a powerful tool for exploring the structure of the DNA helix. Their interactions have been intensively studied because of the excellent photophysical and photochemical properties of ruthenium compounds. In this perspective, this review describes the recent developments in the interactions of these two classic intercalated compounds with a DNA helix. The mechanism of the molecular light switch effect and the selectivity of these two compounds to different forms of a DNA helix has been discussed. In addition, the specific binding modes between them have been discussed in detail, for a better understanding the mechanism of the light switch and the luminescence difference. Finally, recent studies of single molecule force spectroscopy have also been included so as to precisely interpret the kinetics, equilibrium constants, and the energy landscape during the process of the dynamic assembly of ligands into a single DNA helix.
Keywords: binding mode; intercalated ruthenium compounds; molecular light switch; sensitive luminescent reporter; single molecule force spectroscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
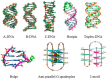

Similar articles
-
Enantioselective luminescence quenching of DNA light-switch [Ru(phen)2dppz]2+ by electron transfer to structural homologue [Ru(phendione)2dppz]2+.J Phys Chem B. 2005 Sep 15;109(36):17327-32. doi: 10.1021/jp0517091. J Phys Chem B. 2005. PMID: 16853212
-
A theoretical study of Ru(II) polypyridyl DNA intercalators structure and electronic absorption spectroscopy of [Ru(phen)2(dppz)]2+ and [Ru(tap)2(dppz)]2+ complexes intercalated in guanine-cytosine base pairs.J Inorg Biochem. 2010 Sep;104(9):893-901. doi: 10.1016/j.jinorgbio.2010.04.002. Epub 2010 Apr 14. J Inorg Biochem. 2010. PMID: 20554006
-
Ruthenium(ii) complexes with dppz: from molecular photoswitch to biological applications.Dalton Trans. 2016 Sep 14;45(34):13261-76. doi: 10.1039/c6dt01624c. Epub 2016 Jul 18. Dalton Trans. 2016. PMID: 27426487 Review.
-
Synthesis, DNA-binding and photocleavage studies of [Ru(phen)2(pbtp)]2+ and [Ru(bpy)2(pbtp)]2+ (phen=1,10-phenanthroline; bpy=2,2'-bipyridine; pbtp=4,5,9,11,14-pentaaza-benzo[b]triphenylene).Spectrochim Acta A Mol Biomol Spectrosc. 2009 Sep 1;73(5):858-64. doi: 10.1016/j.saa.2009.04.021. Epub 2009 May 3. Spectrochim Acta A Mol Biomol Spectrosc. 2009. PMID: 19497781
-
Recent developments in the nanostructured materials functionalized with ruthenium complexes for targeted drug delivery to tumors.Int J Nanomedicine. 2017 Apr 4;12:2749-2758. doi: 10.2147/IJN.S131304. eCollection 2017. Int J Nanomedicine. 2017. PMID: 28435255 Free PMC article. Review.
Cited by
-
A Fluorescent "Turn-On" Clutch Probe for Plasma Cell-Free DNA Identification from Lung Cancer Patients.Nanomaterials (Basel). 2022 Apr 8;12(8):1262. doi: 10.3390/nano12081262. Nanomaterials (Basel). 2022. PMID: 35457970 Free PMC article.
-
Computational Assessment of a Dual-Action Ru(II)-Based Complex: Photosensitizer in Photodynamic Therapy and Intercalating Agent for Inducing DNA Damage.Inorg Chem. 2023 Jun 12;62(23):8948-8959. doi: 10.1021/acs.inorgchem.3c00592. Epub 2023 May 29. Inorg Chem. 2023. PMID: 37248070 Free PMC article.
-
Photocytotoxic Activity of Ruthenium(II) Complexes with Phenanthroline-Hydrazone Ligands.Molecules. 2021 Apr 6;26(7):2084. doi: 10.3390/molecules26072084. Molecules. 2021. PMID: 33917290 Free PMC article.
-
Metal-Based Drug-DNA Interactions and Analytical Determination Methods.Molecules. 2024 Sep 13;29(18):4361. doi: 10.3390/molecules29184361. Molecules. 2024. PMID: 39339356 Free PMC article. Review.
References
-
- Belmont P., Constant J.F., Demeunynck M. Nucleic Acid Conformation Diversity: From Structure to Function and Regulation. Chem. Soc. Rev. 2001;30:70–81. doi: 10.1039/a904630e. - DOI
-
- Martin E. Nucleic acid crystallography: Current progress. Curr. Opin. Chem. Biol. 2004;8:580–591. - PubMed
-
- Shi S., Yao T., Ji L.N. DNA molecular light switch and biosensors based on ruthenium(II) polypyridyl complexes. Sci. China-Chem. 2014;44:460–470.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

