Use of isotopically labeled substrates reveals kinetic differences between human and bacterial serine palmitoyltransferase
- PMID: 30792183
- PMCID: PMC6495160
- DOI: 10.1194/jlr.M089367
Use of isotopically labeled substrates reveals kinetic differences between human and bacterial serine palmitoyltransferase
Erratum in
-
Erratum: Use of isotopically labeled substrates reveals kinetic differences between human and bacterial serine palmitoyltransferase.J Lipid Res. 2019 Aug;60(8):1489. doi: 10.1194/jlr.ERR119000228. J Lipid Res. 2019. PMID: 31371643 Free PMC article. No abstract available.
Abstract
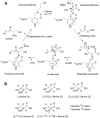
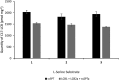
Isotope labels are frequently used tools to track metabolites through complex biochemical pathways and to discern the mechanisms of enzyme-catalyzed reactions. Isotopically labeled l-serine is often used to monitor the activity of the first enzyme in sphingolipid biosynthesis, serine palmitoyltransferase (SPT), as well as labeling downstream cellular metabolites. Intrigued by the effect that isotope labels may be having on SPT catalysis, we characterized the impact of different l-serine isotopologues on the catalytic activity of recombinant SPT isozymes from humans and the bacterium Sphingomonas paucimobilis Our data show that S. paucimobilis SPT activity displays a clear isotope effect with [2,3,3-D]l-serine, whereas the human SPT isoform does not. This suggests that although both human and S. paucimobilis SPT catalyze the same chemical reaction, there may well be underlying subtle differences in their catalytic mechanisms. Our results suggest that it is the activating small subunits of human SPT that play a key role in these mechanistic variations. This study also highlights that it is important to consider the type and location of isotope labels on a substrate when they are to be used in in vitro and in vivo studies.
Keywords: biosynthesis; mechanism; membrane protein; regulation; sphingolipid.
Copyright © 2019 Harrison et al.
Figures

Similar articles
-
The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics.J Biol Chem. 2009 Jun 19;284(25):17328-17339. doi: 10.1074/jbc.M109.008680. Epub 2009 Apr 17. J Biol Chem. 2009. PMID: 19376777 Free PMC article.
-
Reconstitution of the pyridoxal 5'-phosphate (PLP) dependent enzyme serine palmitoyltransferase (SPT) with pyridoxal reveals a crucial role for the phosphate during catalysis.Chem Commun (Camb). 2013 Aug 14;49(63):7058-60. doi: 10.1039/c3cc43001d. Chem Commun (Camb). 2013. PMID: 23814788
-
The pyridoxal 5'-phosphate (PLP)-dependent enzyme serine palmitoyltransferase (SPT): effects of the small subunits and insights from bacterial mimics of human hLCB2a HSAN1 mutations.Biomed Res Int. 2013;2013:194371. doi: 10.1155/2013/194371. Epub 2013 Sep 23. Biomed Res Int. 2013. PMID: 24175284 Free PMC article.
-
Bacterial serine palmitoyltransferase: a water-soluble homodimeric prototype of the eukaryotic enzyme.Biochim Biophys Acta. 2003 Apr 11;1647(1-2):116-20. doi: 10.1016/s1570-9639(03)00074-8. Biochim Biophys Acta. 2003. PMID: 12686119 Review.
-
Structural, mechanistic and regulatory studies of serine palmitoyltransferase.Biochem Soc Trans. 2012 Jun 1;40(3):547-54. doi: 10.1042/BST20110769. Biochem Soc Trans. 2012. PMID: 22616865 Review.
Cited by
-
Convergent evolution of bacterial ceramide synthesis.Nat Chem Biol. 2022 Mar;18(3):305-312. doi: 10.1038/s41589-021-00948-7. Epub 2021 Dec 30. Nat Chem Biol. 2022. PMID: 34969973 Free PMC article.
-
Identification of a novel SPT inhibitor WXP-003 by docking-based virtual screening and investigation of its anti-fungi effect.J Enzyme Inhib Med Chem. 2021 Dec;36(1):1007-1015. doi: 10.1080/14756366.2021.1915301. J Enzyme Inhib Med Chem. 2021. PMID: 34148472 Free PMC article.
-
Don't Be Surprised When These Surprise You: Some Infrequently Studied Sphingoid Bases, Metabolites, and Factors That Should Be Kept in Mind During Sphingolipidomic Studies.Int J Mol Sci. 2025 Jan 14;26(2):650. doi: 10.3390/ijms26020650. Int J Mol Sci. 2025. PMID: 39859363 Free PMC article. Review.
-
Desaturation of the Sphingofungin Polyketide Tail Results in Increased Serine Palmitoyltransferase Inhibition.Microbiol Spectr. 2022 Oct 26;10(5):e0133122. doi: 10.1128/spectrum.01331-22. Epub 2022 Sep 19. Microbiol Spectr. 2022. PMID: 36121228 Free PMC article.
-
Porphyromonas gingivalis Sphingolipid Synthesis Limits the Host Inflammatory Response.J Dent Res. 2020 May;99(5):568-576. doi: 10.1177/0022034520908784. Epub 2020 Feb 27. J Dent Res. 2020. PMID: 32105543 Free PMC article.
References
-
- Lowther J., Naismith J. H., Dunn T. M., and Campopiano D. J.. 2012. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40: 547–554. - PubMed
-
- Yard B. A., Carter L. G., Johnson K. A., Overton I. M., Dorward M., Liu H., McMahon S. A., Oke M., Puech D., Barton G. J., et al. . 2007. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J. Mol. Biol. 370: 870–886. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

