Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients
- PMID: 30792206
- PMCID: PMC6886416
- DOI: 10.3324/haematol.2018.207241
Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients
Erratum in
-
Pathogenic mutations identified by a multimodality approach in 117 Japanese Fanconi anemia patients.Haematologica. 2020 Apr;105(4):1166-1167. doi: 10.3324/haematol.2019.245720. Haematologica. 2020. PMID: 32238468 Free PMC article. No abstract available.
Abstract
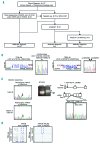
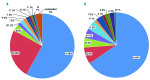
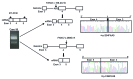
Fanconi anemia is a rare recessive disease characterized by multiple congenital abnormalities, progressive bone marrow failure, and a predisposition to malignancies. It results from mutations in one of the 22 known FANC genes. The number of Japanese Fanconi anemia patients with a defined genetic diagnosis was relatively limited. In this study, we reveal the genetic subtyping and the characteristics of mutated FANC genes in Japan and clarify the genotype-phenotype correlations. We studied 117 Japanese patients and successfully subtyped 97% of the cases. FANCA and FANCG pathogenic variants accounted for the disease in 58% and 25% of Fanconi anemia patients, respectively. We identified one FANCA and two FANCG hot spot mutations, which are found at low percentages (0.04-0.1%) in the whole-genome reference panel of 3,554 Japanese individuals (Tohoku Medical Megabank). FANCB was the third most common complementation group and only one FANCC case was identified in our series. Based on the data from the Tohoku Medical Megabank, we estimate that approximately 2.6% of Japanese are carriers of disease-causing FANC gene variants, excluding missense mutations. This is the largest series of subtyped Japanese Fanconi anemia patients to date and the results will be useful for future clinical management.
Copyright© 2019 Ferrata Storti Foundation.
Figures
References
-
- Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 2018;18(3):168-185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

