Hachimoji DNA and RNA: A genetic system with eight building blocks
- PMID: 30792304
- PMCID: PMC6413494
- DOI: 10.1126/science.aat0971
Hachimoji DNA and RNA: A genetic system with eight building blocks
Abstract
We report DNA- and RNA-like systems built from eight nucleotide "letters" (hence the name "hachimoji") that form four orthogonal pairs. These synthetic systems meet the structural requirements needed to support Darwinian evolution, including a polyelectrolyte backbone, predictable thermodynamic stability, and stereoregular building blocks that fit a Schrödinger aperiodic crystal. Measured thermodynamic parameters predict the stability of hachimoji duplexes, allowing hachimoji DNA to increase the information density of natural terran DNA. Three crystal structures show that the synthetic building blocks do not perturb the aperiodic crystal seen in the DNA double helix. Hachimoji DNA was then transcribed to give hachimoji RNA in the form of a functioning fluorescent hachimoji aptamer. These results expand the scope of molecular structures that might support life, including life throughout the cosmos.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

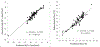


Comment in
-
Four new DNA letters double life's alphabet.Nature. 2019 Feb;566(7745):436. doi: 10.1038/d41586-019-00650-8. Nature. 2019. PMID: 30809059 No abstract available.
-
Eight-Letter DNA.Biochemistry. 2019 Jun 4;58(22):2581-2583. doi: 10.1021/acs.biochem.9b00274. Epub 2019 May 22. Biochemistry. 2019. PMID: 31117391 Free PMC article.
References
-
- Watson JD, Crick FHC, Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953). - PubMed
-
- Schrödinger E, Was ist Leben Serie Piper, Vol. 1134 (1943).
-
- Goodman MF, On the wagon. DNA polymerase joins “H-bonds anonymous”. Nature Biotechnol 17, 640–640 (1999). - PubMed
-
- Kimoto M, Yamashige R, Matsunaga KI, Yokoyama S, Hirao I, Generation of high- affinity DNA aptamers using an expanded genetic alphabet. Nature Biotechnol 31, 453–457 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

