Functional reconstitution of vacuolar H+-ATPase from Vo proton channel and mutant V1-ATPase provides insight into the mechanism of reversible disassembly
- PMID: 30792311
- PMCID: PMC6484122
- DOI: 10.1074/jbc.RA119.007577
Functional reconstitution of vacuolar H+-ATPase from Vo proton channel and mutant V1-ATPase provides insight into the mechanism of reversible disassembly
Abstract
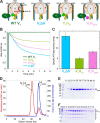
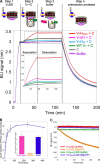
The vacuolar H+-ATPase (V-ATPase; V1Vo-ATPase) is an ATP-dependent proton pump that acidifies subcellular compartments in all eukaryotic organisms. V-ATPase activity is regulated by reversible disassembly into autoinhibited V1-ATPase and Vo proton channel subcomplexes, a process that is poorly understood on the molecular level. V-ATPase is a rotary motor, and recent structural analyses have revealed different rotary states for disassembled V1 and Vo, a mismatch that is likely responsible for their inability to reconstitute into holo V-ATPase in vitro Here, using the model organism Saccharomyces cerevisiae, we show that a key impediment for binding of V1 to Vo is the conformation of the inhibitory C-terminal domain of subunit H (HCT). Using biolayer interferometry and biochemical analyses of purified mutant V1-ATPase and Vo proton channel reconstituted into vacuolar lipid-containing nanodiscs, we further demonstrate that disruption of HCT's V1-binding site facilitates assembly of a functionally coupled and stable V1Vo-ATPase. Unlike WT, this mutant enzyme was resistant to MgATP hydrolysis-induced dissociation, further highlighting HCT's role in the mechanism of V-ATPase regulation. Our findings provide key insight into the molecular events underlying regulation of V-ATPase activity by reversible disassembly.
Keywords: biolayer interferometry; biophysics; lipid nanodisc; membrane protein; molecular motor; protein-protein interaction; reversible disassembly; vacuolar ATPase.
© 2019 Sharma et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Karet F. E., Finberg K. E., Nelson R. D., Nayir A., Mocan H., Sanjad S. A., Rodriguez-Soriano J., Santos F., Cremers C. W., Di Pietro A., Hoffbrand B. I., Winiarski J., Bakkaloglu A., Ozen S., Dusunsel R., et al. (1999) Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat. Genet. 21, 84–90 10.1038/5022 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

