Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D
- PMID: 30792383
- PMCID: PMC6385193
- DOI: 10.1038/s41419-019-1399-2
Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D
Abstract
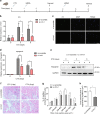
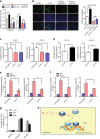
Recent studies suggest important roles for long non-coding RNAs as essential regulators of myogenic differentiation. Here, we report that lncRNA Irm is upregulated during myogenesis. Functional analyses show that the overexpression of Irm enhances myogenic differentiation, whereas the inhibition of Irm has completely opposite effects in vitro. Notably, the inhibition of Irm blocks damage-induced muscle regeneration in vivo. Mechanistically, Irm regulates the expression of myogenic genes by directly binding to MEF2D, which in turn promotes the assembly of MyoD/MEF2D on the regulatory elements of target genes. Collectively, we have identified a novel lncRNA that interacts with MEF2D to regulate myogenesis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

