Extreme slow growth as alternative strategy to survive deep starvation in bacteria
- PMID: 30792386
- PMCID: PMC6385201
- DOI: 10.1038/s41467-019-08719-8
Extreme slow growth as alternative strategy to survive deep starvation in bacteria
Abstract
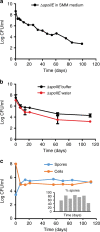
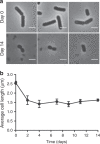
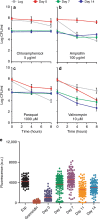
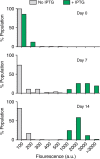
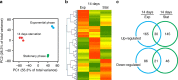
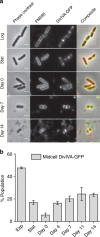
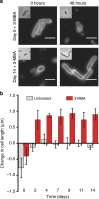
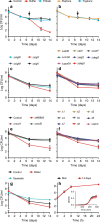
Bacteria can become dormant or form spores when they are starved for nutrients. Here, we find that non-sporulating Bacillus subtilis cells can survive deep starvation conditions for many months. During this period, cells adopt an almost coccoid shape and become tolerant to antibiotics. Unexpectedly, these cells appear to be metabolically active and show a transcriptome profile very different from that of stationary phase cells. We show that these starved cells are not dormant but are growing and dividing, albeit with a doubling time close to 4 days. Very low nutrient levels, comparable to 10,000-fold diluted lysogeny broth (LB), are sufficient to sustain this growth. This extreme slow growth, which we propose to call 'oligotrophic growth state', provides an alternative strategy for B. subtilis to endure nutrient depletion and environmental stresses. Further work is warranted to test whether this state can be found in other bacterial species to survive deep starvation conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
Life in the slow lane.Nat Rev Microbiol. 2019 May;17(5):266-267. doi: 10.1038/s41579-019-0176-7. Nat Rev Microbiol. 2019. PMID: 30833717 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

