A systematic study on the influence of thermodynamic asymmetry of 5'-ends of siRNA duplexes in relation to their silencing potency
- PMID: 30792489
- PMCID: PMC6385221
- DOI: 10.1038/s41598-018-36620-9
A systematic study on the influence of thermodynamic asymmetry of 5'-ends of siRNA duplexes in relation to their silencing potency
Abstract
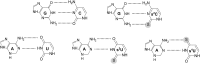
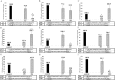
siRNA molecules possess high potential as molecular tools and can be used as effective therapeutics in humans. One of the key steps in the action of these molecules is the choice of antisense strand by the RNA-induced silencing complex (RISC). To explain this process, we verified the theory which states that antisense strand selection is based on the thermodynamically less stable 5' end of siRNA. Based on the studies presented herein, we observed that for the tested siRNA duplexes, the difference in the thermodynamic stability of the terminal, penultimate and pre-penultimate pairs in the duplex siRNA is not the dominant factor in antisense strand selection. We found that both strands in each tested siRNA molecule are used as an antisense strand. The introduction of modified nucleotides, whose impact on the thermodynamic stability of siRNA duplexes was studied, results in changes in antisense strand selection by the RISC complex. The presence of a modified residue often caused predominant selection of only one antisense strand which is at variance with the theory of siRNA strand bias.
Conflict of interest statement
The authors declare no competing interests.
Figures

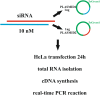
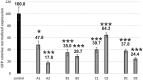
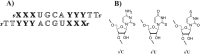
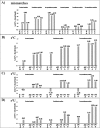

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

