Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico
- PMID: 30792901
- PMCID: PMC6375919
- DOI: 10.1038/s41525-018-0076-1
Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico
Abstract
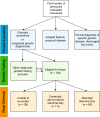
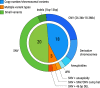
Patients with rare, undiagnosed, or genetic disease (RUGD) often undergo years of serial testing, commonly referred to as the "diagnostic odyssey". Patients in resource-limited areas face even greater challenges-a definitive diagnosis may never be reached due to difficulties in gaining access to clinicians, appropriate specialists, and diagnostic testing. Here, we report on a collaboration of the Illumina iHope Program with the Foundation for the Children of the Californias and Hospital Infantil de Las Californias, to enable deployment of clinical whole genome sequencing (cWGS) as first-tier test in a resource-limited dysmorphology clinic in northern Mexico. A total of 60 probands who were followed for a suspected genetic diagnosis and clinically unresolved after expert examination were tested with cWGS, and the ordering clinicians completed a semi-structured survey to investigate change in clinical management resulting from cWGS findings. Clinically significant genomic findings were identified in 68.3% (n = 41) of probands. No recurrent molecular diagnoses were observed. Copy number variants or gross chromosomal abnormalities accounted for 48.8% (n = 20) of the diagnosed cases, including a mosaic trisomy and suspected derivative chromosomes. A qualitative assessment of clinical management revealed 48.8% (n = 20) of those diagnosed had a change in clinical course based on their cWGS results, despite resource limitations. These data suggest that a cWGS first-tier testing approach can benefit patients with suspected genetic disorders.
Conflict of interest statement
A.S., A.G., J.E., K.R., V.R., E.T., S.S.A., D.L.P., D.R.B., J.W.B., R.J.T., and all members of the ICSL interpretation and reporting team are employees and shareholders of Illumina Inc. The remaining authors declare no competing interests.
Figures
References
-
- Shire. Rare disease impact report: Insights from patients and the medical community. https://globalgenes.org/wp-content/uploads/2013/04/ShireReport-1.pdf (2013).
-
- Modell B, Kuliev A. The history of community genetics: the contribution of the haemoglobin disorders. Community Genet. 1998;1:3–11. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

