Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses
- PMID: 30792902
- PMCID: PMC6371749
- DOI: 10.1093/ve/vey040
Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses
Abstract
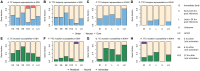
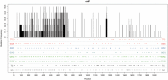
The human gamma-herpesviruses, Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus, establish lifelong latency in B cells and are associated with multiple malignancies. Virus-host coevolution often drive changes in both host immunity and in the viral genome. We consider one host immune mechanism, the activation-induced deaminase (AID)/APOBEC family of cytidine deaminases, that induces mutations in viral DNA. AID, the ancestral gene in the family has a conserved role in somatic hypermutation, a key step in antibody affinity maturation. The APOBEC3 subfamily, of which there are seven genes in human, have evolved antiviral functions and have diversified in terms of their expression pattern, subcellular localization, and DNA mutation motifs (hotspots). In this study, we investigated how the human gamma-herpesviruses have evolved to avoid the action of the AID/APOBEC enzymes and determine if these enzymes are contributing to the ongoing evolution of the viruses. We used computational methods to evaluate observed versus expected frequency of AID/APOBEC hotspots in viral genomes and found that the viruses have evolved to limit the representation of AID and certain APOBEC3 motifs. At the same time, the remaining hotspots were highly likely to cause amino acid changes, suggesting prolonged evolutionary pressure of the enzymes on the viruses. To study current hypermutation, as opposed to historical mutation processes, we also analyzed putative mutations derived from alignments of published viral genomes and found again that AID and APOBEC3 appear to target the genome most frequently. New protein variants resulting from AID/APOBEC activity may have important consequences in health, including vaccine development (epitope evolution) and host immune evasion.
Keywords: AID/APOBEC; evolution; gamma-herpesviruses.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources

