A modified protocol for rapid DNA isolation from cotton (Gossypium spp.)
- PMID: 30792967
- PMCID: PMC6370549
- DOI: 10.1016/j.mex.2019.01.010
A modified protocol for rapid DNA isolation from cotton (Gossypium spp.)
Abstract
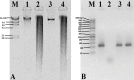
Extraction of high-quality DNA from Gossypium (Cotton) species is notoriously difficult due to high contents of polysaccharides, quinones and polyphenols other secondary metabolites. Here, we describe a simple, rapid and modified procedure for high-quality DNA extraction from cotton, which is amenable for downstream analyses. In contrast to other CTAB methods, the described procedure is rapid, omits the use of liquid nitrogen, phenol, CsCl gradient ultracentrifugation, uses inexpensive and less hazardous reagents, and requires only ordinary laboratory equipment. The procedure employed the high concentration of NaCl and use of PVP-10 to rid the problems associated with polysaccharides and polyphenols, respectively. The average yield was approximately 10-15 μg of good quality DNA from 100 mg of tissue weight, which is adequate for projects, like genetic mapping or marker-assisted plant breeding. This protocol can be performed in as little as 3 h and may be adapted to high-throughput DNA isolation. •Buffers A and B were redesigned from Paterson et al. (1993) and Porebski et al. (1997), respectively.•Ribonuclease A was added before chloroform extraction.•A simple, rapid and inexpensive DNA extraction method is described.
Keywords: CTAB; CTAB, cetyltrimethylammonium bromide; DNA extraction; Gossypium; PVP-10, polyvinylpyrrolidone; Polyphenols; Polysaccharides; Without liquid-nitrogen.
Figures
Similar articles
-
A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide.Nat Protoc. 2006;1(5):2320-5. doi: 10.1038/nprot.2006.384. Nat Protoc. 2006. PMID: 17406474
-
An improved CTAB-ammonium acetate method for total RNA isolation from cotton.Phytochem Anal. 2012 Nov-Dec;23(6):647-50. doi: 10.1002/pca.2368. Epub 2012 May 3. Phytochem Anal. 2012. PMID: 22552877
-
Cationic and anionic detergent buffers in sequence yield high-quality genomic DNA from diverse plant species.Anal Biochem. 2024 Jan 1;684:115372. doi: 10.1016/j.ab.2023.115372. Epub 2023 Nov 7. Anal Biochem. 2024. PMID: 37940013
-
DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol.ISRN Mol Biol. 2012 Nov 14;2012:205049. doi: 10.5402/2012/205049. eCollection 2012. ISRN Mol Biol. 2012. PMID: 27335662 Free PMC article.
-
An improved Shorea robusta genomic DNA extraction protocol with high PCR fidelity.Biol Methods Protoc. 2023 Dec 9;8(1):bpad039. doi: 10.1093/biomethods/bpad039. eCollection 2023. Biol Methods Protoc. 2023. PMID: 38116323 Free PMC article. Review.
Cited by
-
An SNP based genotyping assay for genes associated with drought tolerance in bread wheat.Mol Biol Rep. 2024 Apr 18;51(1):527. doi: 10.1007/s11033-024-09481-x. Mol Biol Rep. 2024. PMID: 38637351
-
A single-nucleotide mutation of G301A in GaIAA14 confers leaf curling in Gossypium arboreum.Front Plant Sci. 2025 Jul 22;16:1645239. doi: 10.3389/fpls.2025.1645239. eCollection 2025. Front Plant Sci. 2025. PMID: 40765858 Free PMC article.
-
Research Progress of Nucleic Acid Detection Technology for Genetically Modified Maize.Int J Mol Sci. 2023 Jul 31;24(15):12247. doi: 10.3390/ijms241512247. Int J Mol Sci. 2023. PMID: 37569623 Free PMC article. Review.
-
New Insights into Methyl Jasmonate Regulation of Triterpenoid Biosynthesis in Medicinal Fungal Species Sanghuangporusbaumii (Pilát) L.W. Zhou & Y.C. Dai.J Fungi (Basel). 2022 Aug 23;8(9):889. doi: 10.3390/jof8090889. J Fungi (Basel). 2022. PMID: 36135614 Free PMC article.
-
Functional characterization of GhNAC2 promoter conferring hormone- and stress-induced expression: a potential tool to improve growth and stress tolerance in cotton.Physiol Mol Biol Plants. 2024 Jan;30(1):17-32. doi: 10.1007/s12298-024-01411-2. Epub 2024 Feb 8. Physiol Mol Biol Plants. 2024. PMID: 38435854 Free PMC article.
References
-
- Varma A., Padh H., Shrivastava N. Plant genomic DNA isolation: an art or a science. Biotechnol. J. 2007;2:386–392. - PubMed
-
- Kalbande B.B., Patil A.S., Chakrabarty P.K. An efficient, simple and high throughput protocol for cotton genomic DNA isolation. J. Plant Biochem. Biotechnol. 2016;25:437–441.
-
- Arruda S.R., Pereira D.G., Silva-Castro M.M., Brito M.G., Waldschmidt A.M. An optimized protocol for DNA extraction in plants with a high content of secondary metabolites, based on leaves of Mimosa tenuiflora (Willd.) Poir. (Leguminosae) Genet. Mol. Res. 2017;16:3. - PubMed
-
- Porebski S., Bailey L.G., Baum B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997;15:8–15.
LinkOut - more resources
Full Text Sources
Miscellaneous

