The modular organization of human anatomical brain networks: Accounting for the cost of wiring
- PMID: 30793069
- PMCID: PMC6372290
- DOI: 10.1162/NETN_a_00002
The modular organization of human anatomical brain networks: Accounting for the cost of wiring
Abstract
Brain networks are expected to be modular. However, existing techniques for estimating a network's modules make it difficult to assess the influence of organizational principles such as wiring cost reduction on the detected modules. Here we present a modification of an existing module detection algorithm that allowed us to focus on connections that are unexpected under a cost-reduction wiring rule and to identify modules from among these connections. We applied this technique to anatomical brain networks and showed that the modules we detected differ from those detected using the standard technique. We demonstrated that these novel modules are spatially distributed, exhibit unique functional fingerprints, and overlap considerably with rich clubs, giving rise to an alternative and complementary interpretation of the functional roles of specific brain regions. Finally, we demonstrated that, using the modified module detection approach, we can detect modules in a developmental dataset that track normative patterns of maturation. Collectively, these findings support the hypothesis that brain networks are composed of modules and provide additional insight into the function of those modules.
Keywords: Community structure; Complex networks; Geometry; Modularity; Wiring cost.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures





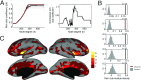
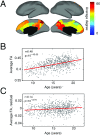
References
-
- Ahn Y. Y., Bagrow J. P., & Lehmann S. (2010). Link communities reveal multiscale complexity in networks. Nature, 466(7307), 761– 764. - PubMed
-
- Arenas A., Fernandez A., & Gomez S. (2008). Analysis of the structure of complex networks at different resolution levels. New Journal of Physics, 10(5), 053039.
Grants and funding
LinkOut - more resources
Full Text Sources
