Computation is concentrated in rich clubs of local cortical networks
- PMID: 30793088
- PMCID: PMC6370472
- DOI: 10.1162/netn_a_00069
Computation is concentrated in rich clubs of local cortical networks
Abstract
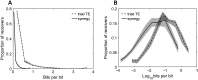
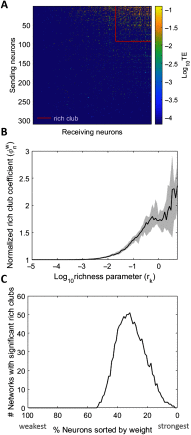
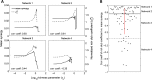
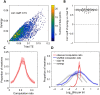
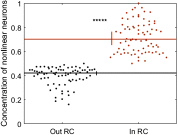
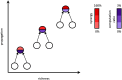
To understand how neural circuits process information, it is essential to identify the relationship between computation and circuit organization. Rich clubs, highly interconnected sets of neurons, are known to propagate a disproportionate amount of information within cortical circuits. Here, we test the hypothesis that rich clubs also perform a disproportionate amount of computation. To do so, we recorded the spiking activity of on average ∼300 well-isolated individual neurons from organotypic cortical cultures. We then constructed weighted, directed networks reflecting the effective connectivity between the neurons. For each neuron, we quantified the amount of computation it performed based on its inputs. We found that rich-club neurons compute ∼160% more information than neurons outside of the rich club. The amount of computation performed in the rich club was proportional to the amount of information propagation by the same neurons. This suggests that in these circuits, information propagation drives computation. In total, our findings indicate that rich-club organization in effective cortical circuits supports not only information propagation but also neural computation.
Keywords: Cortical networks; Effective connectivity; Information theory; Neural computation; Rich clubs; Transfer entropy.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures




References
-
- Adamatzky A., & Durand-Lose J. (2012). Collision-based computing In Rozenberg G., Bäck T., & Kok J. N. (Eds.), Handbook of Natural Computing (pp. 1949–1978). Berlin: Springer.
-
- Bertschinger N., Rauh J., Olbrich E., Jost J., & Ay N. (2014). Quantifying unique information. Entropy, 16(4), 2161–2183.
-
- Bhattacharjee K., Naskar N., Roy S., & Das S. (2016). A survey of cellular automata: Types, dynamics, non-uniformity and applications. arXiv:1607.02291.
-
- Bolz J., Novak N., Götz M., & Bonhoeffer T. (1990). Formation of target-specific neuronal projections in organotypic slice cultures from rat visual cortex. Nature, 346, 359–362. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
