Analysis of the Prader-Willi syndrome imprinting center using droplet digital PCR and next-generation whole-exome sequencing
- PMID: 30793526
- PMCID: PMC6465664
- DOI: 10.1002/mgg3.575
Analysis of the Prader-Willi syndrome imprinting center using droplet digital PCR and next-generation whole-exome sequencing
Abstract
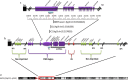
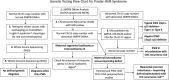
Background: Detailed analysis of imprinting center (IC) defects in individuals with Prader-Willi syndrome (PWS) is not readily available beyond chromosomal microarray (MA) analysis, and such testing is important for a more accurate diagnosis and recurrence risks. This is the first feasibility study of newly developed droplet digital polymerase chain reaction (ddPCR) examining DNA copy number differences in the PWS IC region of those with IC defects.
Methods: The study cohort included 17 individuals without 15q11-q13 deletions or maternal disomy but with IC defects as determined by genotype analysis showing biparental inheritance. Seven sets of parents and two healthy, unrelated controls were also analyzed.
Results: Copy number differences were distinguished by comparing the number of positive droplets detected by IC probes to those from a chromosome 15 reference probe, GABRβ3. The ddPCR findings were compared to results from other methods including MA, and whole-exome sequencing (WES) with 100% concordance. The study also estimated the frequency of IC microdeletions and identified gene variants by WES that may impact phenotypes including CPT2 and NTRK1 genes.
Conclusion: Droplet digital polymerase chain reaction is a cost-effective method that can be used to confirm the presence of microdeletions in PWS with impact on genetic counseling and recurrence risks for families.
Keywords: Prader-Willi syndrome (PWS); droplet digital PCR; epimutation; imprinting center; microdeletion; whole-exome sequencing.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
None declared.
Figures
Similar articles
-
Influence of the Prader-Willi syndrome imprinting center on the DNA methylation landscape in the mouse brain.Epigenetics. 2014 Nov;9(11):1540-56. doi: 10.4161/15592294.2014.969667. Epigenetics. 2014. PMID: 25482058 Free PMC article.
-
[Prader-Willi syndrome and genomic imprinting].Zhonghua Er Ke Za Zhi. 2003 Jun;41(6):453-6. Zhonghua Er Ke Za Zhi. 2003. PMID: 14749005 Chinese.
-
Three siblings with Prader-Willi syndrome caused by imprinting center microdeletions and review.Am J Med Genet A. 2018 Apr;176(4):886-895. doi: 10.1002/ajmg.a.38627. Epub 2018 Feb 13. Am J Med Genet A. 2018. PMID: 29437285 Free PMC article.
-
Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes.Mol Hum Reprod. 1997 Apr;3(4):321-32. doi: 10.1093/molehr/3.4.321. Mol Hum Reprod. 1997. PMID: 9237260 Review.
-
Prader-Willi syndrome: reflections on seminal studies and future therapies.Open Biol. 2020 Sep;10(9):200195. doi: 10.1098/rsob.200195. Epub 2020 Sep 23. Open Biol. 2020. PMID: 32961075 Free PMC article. Review.
Cited by
-
Early Diagnosis in Prader-Willi Syndrome Reduces Obesity and Associated Co-Morbidities.Genes (Basel). 2019 Nov 6;10(11):898. doi: 10.3390/genes10110898. Genes (Basel). 2019. PMID: 31698873 Free PMC article.
-
Chromosome 15 Imprinting Disorders: Genetic Laboratory Methodology and Approaches.Front Pediatr. 2020 May 12;8:154. doi: 10.3389/fped.2020.00154. eCollection 2020. Front Pediatr. 2020. PMID: 32478012 Free PMC article. Review.
-
Genotype-Phenotype Relationships and Endocrine Findings in Prader-Willi Syndrome.Front Endocrinol (Lausanne). 2019 Dec 13;10:864. doi: 10.3389/fendo.2019.00864. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31920975 Free PMC article. Review.
-
Recommendations for the diagnosis and management of childhood Prader-Willi syndrome in China.Orphanet J Rare Dis. 2022 Jun 13;17(1):221. doi: 10.1186/s13023-022-02302-z. Orphanet J Rare Dis. 2022. PMID: 35698200 Free PMC article. Review.
-
Clinical Assessment, Genetics, and Treatment Approaches in Autism Spectrum Disorder (ASD).Int J Mol Sci. 2020 Jul 2;21(13):4726. doi: 10.3390/ijms21134726. Int J Mol Sci. 2020. PMID: 32630718 Free PMC article. Review.
References
-
- Buiting, K. , Gross, S. , Lich, C. , Gillessen‐Kaesbach, G. , El‐Maarri, O. , & Horsthemke, B. (2003). Epimutations in Prader‐Willi and Angelman syndromes: A molecular study of 136 patients with an imprinting defect. The American Journal of Human Genetics, 72(3), 571–577. 10.1086/367926 - DOI - PMC - PubMed
-
- Buiting, K. , Färbe, R. C. , Kroisel, P. , Wagner, K. , Brueton, L. , Robertson, M. E. , … Horsthemke, B. (2000). Imprinting centre deletions in two PWS families: Implications for diagnostic testing and genetic counselling. Clinical Genetics, 58(4), 284–290. 10.1034/j.1399-0004.2000.580406.x - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

