The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: Evidence from bioinformatics analysis of high-throughput data
- PMID: 30793530
- PMCID: PMC6503072
- DOI: 10.1002/mgg3.607
The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: Evidence from bioinformatics analysis of high-throughput data
Abstract
Background: Kidney renal clear cell carcinoma (KIRC) is the most common subtype of renal tumor. However, the molecular mechanisms of KIRC pathogenesis remain little known. The purpose of our study was to identify potential key genes related to the occurrence and prognosis of KIRC, which could serve as novel diagnostic and prognostic biomarkers for KIRC.
Methods: Three gene expression profiles from gene expression omnibus database were integrated to identify differential expressed genes (DEGs) using limma package. Enrichment analysis and PPI construction for these DEGs were performed by bioinformatics tools. We used Gene Expression Profiling Interactive Analysis (GEPIA) database to further analyze the expression and prognostic values of hub genes. The GEPIA database was used to further validate the bioinformatics results. The Connectivity Map was used to identify candidate small molecules that could reverse the gene expression of KIRC.
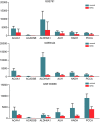
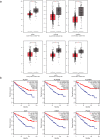
Results: A total of 503 DEGs were obtained. The PPI network with 417 nodes and 1912 interactions was constructed. Go and KEGG pathway analysis revealed that these DEGs were most significantly enriched in excretion and valine, leucine, and isoleucine degradation, respectively. Six DEGs with high degree of connectivity (ACAA1, ACADSB, ALDH6A1, AUH, HADH, and PCCA) were selected as hub genes, which significantly associated with worse survival of patients. Finally, we identified the top 20 most significant small molecules and pipemidic acid was the most promising small molecule to reverse the KIRC gene expression.
Conclusions: This study first uncovered six key genes in KIRC which contributed to improving our understanding of the molecular mechanisms of KIRC pathogenesis. ACAA1, ACADSB, ALDH6A1, AUH, HADH, and PCCA could serve as the promising novel biomarkers for KIRC diagnosis, prognosis, and treatment.
Keywords: bioinformatics analysis; candidate small molecules; kidney renal clear cell carcinoma; novel biomarkers.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
None.
Figures







References
-
- Bandettini, W. P. , Kellman, P. , Mancini, C. , Booker, O. J. , Vasu, S. , Leung, S. W. , … Arai, A. E. (2012). MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. Journal of Cardiovascular Magnetic Resonance, 14, 83 10.1186/1532-429x-14-83 - DOI - PMC - PubMed
-
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. 10.3322/caac.21492 - DOI - PubMed
-
- Cerami, E. , Gao, J. , Dogrusoz, U. , Gross, B. E. , Sumer, S. O. , Aksoy, B. A. , … Schultz, N. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2(5), 401–404. 10.1158/2159-8290.cd-12-0095 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

