KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer
- PMID: 30794031
- PMCID: PMC6606042
- DOI: 10.1080/15384047.2019.1579959
KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer
Abstract
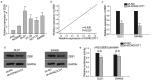
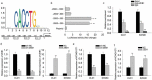
Long noncoding RNAs are widely acknowledged as a group of regulatory factors in various diseases, especially in cancers. KCNQ1 overlapping transcript 1 (KCNQ1OT1) has been reported as oncogene in human cancers. However, the role of KCNQ1OT1 in colorectal cancer (CRC) has not been fully explained. Based on the database analysis, KCNQ1OT1 was highly expressed in CRC samples and predicted the poor prognosis for CRC patients. Functional experiments revealed that KCNQ1OT1 knockdown negatively affected the proliferation, migration and epithelial-mesenchymal transition (EMT) in CRC cells. Moreover, we identified the cytoplasmic localization of KCNQ1OT1 in CRC cells, indicating the post-transcriptional regulation of KCNQ1OT1 on gene expression. Mechanism experiments including RNA Immunoprecipitation (RIP) assay and dual luciferase reporter assays verified that KCNQ1OT1 acted as a competing endogenous RNA (ceRNA) in CRC by sponging microRNA-217 (miR-217) to up-regulate the expression of zinc finger E-box binding homeobox 1 (ZEB1). Further mechanism investigation revealed that ZEB1 enhanced the transcription activity of KCNQ1OT1 by acting as a transcription activator. Finally, rescue assays were designed to demonstrate the effect of KCNQ1OT1-miR-217-ZEB1 feedback loop on proliferation, migration, and EMT of CRC cells. In brief, our research findings revealed that ZEB1-induced upregulation of KCNQ1OT1 improved the proliferation, migration and EMT formation of CRC cells via regulation of miR-217/ZEB1 axis.
Keywords: KCNQ1OT1; ZEB1; colorectal cancer; miR-217; migration.
Figures






References
-
- Du LB, Li HZ, Wang XH, Zhu C, Liu QM, Li QL.. Analysis of cancer incidence in Zhejiang cancer registry in China during 2000 to 2009. Asian Pac J Cancer Prev. 2014;15:5839–5843. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
