A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line
- PMID: 30794539
- PMCID: PMC6402707
- DOI: 10.1371/journal.pgen.1007992
A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line
Abstract
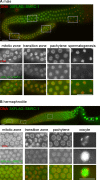
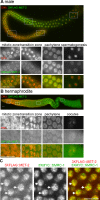
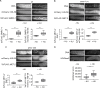
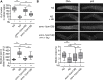
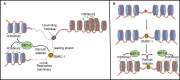
Histone modifications regulate gene expression and chromosomal events, yet how histone-modifying enzymes are targeted is poorly understood. Here we report that a conserved DNA repair protein, SMRC-1, associates with MET-2, the C. elegans histone methyltransferase responsible for H3K9me1 and me2 deposition. We used molecular, genetic, and biochemical methods to investigate the biological role of SMRC-1 and to explore its relationship with MET-2. SMRC-1, like its mammalian ortholog SMARCAL1, provides protection from DNA replication stress. SMRC-1 limits accumulation of DNA damage and promotes germline and embryonic viability. MET-2 and SMRC-1 localize to mitotic and meiotic germline nuclei, and SMRC-1 promotes an increase in MET-2 abundance in mitotic germline nuclei upon replication stress. In the absence of SMRC-1, germline H3K9me2 generally decreases after multiple generations at high culture temperature. Genetic data are consistent with MET-2 and SMRC-1 functioning together to limit replication stress in the germ line and in parallel to promote other germline processes. We hypothesize that loss of SMRC-1 activity causes chronic replication stress, in part because of insufficient recruitment of MET-2 to nuclei.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Madireddy A, Gerhardt J. Replication Through Repetitive DNA Elements and Their Role in Human Diseases In: Masai H, Foiani M, editors. DNA Replication: From Old Principles to New Discoveries. Singapore: Springer Singapore; 2017. p. 549–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

