Identification and characterization of the first pectin methylesterase gene discovered in the root lesion nematode Pratylenchus penetrans
- PMID: 30794636
- PMCID: PMC6386239
- DOI: 10.1371/journal.pone.0212540
Identification and characterization of the first pectin methylesterase gene discovered in the root lesion nematode Pratylenchus penetrans
Erratum in
-
Correction: Identification and characterization of the first pectin methylesterase gene discovered in the root lesion nematode Pratylenchus penetrans.PLoS One. 2019 Mar 18;14(3):e0214185. doi: 10.1371/journal.pone.0214185. eCollection 2019. PLoS One. 2019. PMID: 30883610 Free PMC article.
Abstract
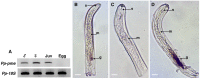
Similar to other plant-parasitic nematodes, root lesion nematodes possess an array of enzymes that are involved in the degradation of the plant cell wall. Here we report the identification of a gene encoding a cell wall-degrading enzyme, pectin methylesterase PME (EC 3.1.1.11), in the root lesion nematode Pratylenchus penetrans. Both genomic and coding sequences of the gene were cloned for this species, that included the presence of four introns which eliminated a possible contamination from bacteria. Expression of the Pp-pme gene was localized in the esophageal glands of P. penetrans as determined by in situ hybridization. Temporal expression of Pp-pme in planta was validated at early time points of infection. The possible function and activity of the gene were assessed by transient expression of Pp-pme in plants of Nicotiana benthamiana plants via a Potato virus X-based vector. To our knowledge, this is the first report on identification and characterization of a PME gene within the phylum Nematoda.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

