Global In-Silico Scenario of tRNA Genes and Their Organization in Virus Genomes
- PMID: 30795514
- PMCID: PMC6409571
- DOI: 10.3390/v11020180
Global In-Silico Scenario of tRNA Genes and Their Organization in Virus Genomes
Abstract
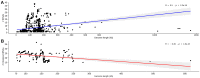
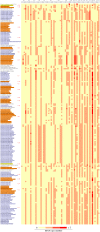
Viruses are known to be highly dependent on the host translation machinery for their protein synthesis. However, tRNA genes are occasionally identified in such organisms, and in addition, few of them harbor tRNA gene clusters comprising dozens of genes. Recently, tRNA gene clusters have been shown to occur among the three domains of life. In such a scenario, the viruses could play a role in the dispersion of such structures among these organisms. Thus, in order to reveal the prevalence of tRNA genes as well as tRNA gene clusters in viruses, we performed an unbiased large-scale genome survey. Interestingly, tRNA genes were predicted in ssDNA (single-stranded DNA) and ssRNA (single-stranded RNA) viruses as well in many other dsDNA viruses of families from Caudovirales order. In the latter group, tRNA gene clusters composed of 15 to 37 tRNA genes were characterized, mainly in bacteriophages, enlarging the occurrence of such structures within viruses. These bacteriophages were from hosts that encompass five phyla and 34 genera. This in-silico study presents the current global scenario of tRNA genes and their organization in virus genomes, contributing and opening questions to be explored in further studies concerning the role of the translation apparatus in these organisms.
Keywords: bacteriophages; codons; host range; ssDNA; ssRNA; tRNA gene; tRNA gene cluster; translation apparatus; virus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Abrahão J., Silva L., Silva L.S., Khalil J.Y.B., Rodrigues R., Arantes T., Assis F., Boratto P., Andrade M., Kroon E.G., et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

