DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae
- PMID: 30795600
- PMCID: PMC6409922
- DOI: 10.3390/genes10020167
DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae
Abstract
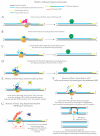
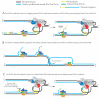
This review discusses a set of experimental results that support the existence of extended strand displacement events during budding yeast lagging strand DNA synthesis. Starting from introducing the mechanisms and factors involved in leading and lagging strand DNA synthesis and some aspects of the architecture of the eukaryotic replisome, we discuss studies on bacterial, bacteriophage and viral DNA polymerases with potent strand displacement activities. We describe proposed pathways of Okazaki fragment processing via short and long flaps, with a focus on experimental results obtained in Saccharomyces cerevisiae that suggest the existence of frequent and extended strand displacement events during eukaryotic lagging strand DNA synthesis, and comment on their implications for genome integrity.
Keywords: DNA helicases; DNA replication; Okazaki fragment processing; flap endonucleases; lagging strand DNA synthesis; strand displacement DNA synthesis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

