Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer
- PMID: 30795624
- PMCID: PMC6406812
- DOI: 10.3390/cancers11020251
Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer
Abstract
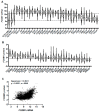
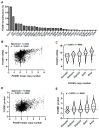
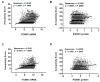
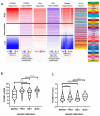
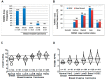
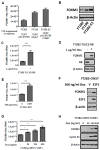
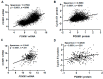
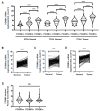
FOXM1 is frequently overexpressed in cancer, but this has not been studied in a comprehensive manner. We utilized genotype-tissue expression (GTEx) normal and The Cancer Genome Atlas (TCGA) tumor data to define FOXM1 expression, including its isoforms, and to determine the genetic alterations that promote FOXM1 expression in cancer. Additionally, we used human fallopian tube epithelial (FTE) cells to dissect the role of Retinoblastoma (Rb)-E2F and Cyclin E1 in FOXM1 regulation, and a novel human embryonic kidney cell (HEK293T) CRISPR FOXM1 knockout model to define isoform-specific transcriptional programs. FOXM1 expression, at the mRNA and protein level, was significantly elevated in tumors with FOXM1 amplification, p53 inactivation, and Rb-E2F deregulation. FOXM1 expression was remarkably high in testicular germ cell tumors (TGCT), high-grade serous ovarian cancer (HGSC), and basal breast cancer (BBC). FOXM1 expression in cancer was associated with genomic instability, as measured using aneuploidy signatures. FTE models confirmed a role for Rb-E2F signaling in FOXM1 regulation and in particular identified Cyclin E1 as a novel inducer of FOXM1 expression. Among the three FOXM1 isoforms, FOXM1c showed the highest expression in normal and tumor tissues and cancer cell lines. The CRISPR knockout model demonstrated that FOXM1b and FOXM1c are transcriptionally active, while FOXM1a is not. Finally, we were unable to confirm the existence of a FOXM1 auto-regulatory loop. This study provides significant and novel information regarding the frequency, causes, and consequences of elevated FOXM1 expression in human cancer.
Keywords: FOXM1; Retinoblastoma protein Cyclin E1; basal breast cancer; fallopian tube epithelial cells; gene amplification; genomic instability; high-grade serous ovarian cancer; pan-cancer; testicular germ cell tumors.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures
References
-
- Ye H., Kelly T.F., Samadani U., Lim L., Rubio S., Overdier D.G., Roebuck K.A., Costa R.H. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 1997;17:1626–1641. doi: 10.1128/MCB.17.3.1626. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

