Influenza Virus Vaccination Elicits Poorly Adapted B Cell Responses in Elderly Individuals
- PMID: 30795982
- PMCID: PMC6452894
- DOI: 10.1016/j.chom.2019.01.002
Influenza Virus Vaccination Elicits Poorly Adapted B Cell Responses in Elderly Individuals
Abstract
Influenza is a leading cause of death in the elderly, and the vaccine protects only a fraction of this population. A key aspect of antibody-mediated anti-influenza virus immunity is adaptation to antigenically distinct epitopes on emerging strains. We examined factors contributing to reduced influenza vaccine efficacy in the elderly and uncovered a dramatic reduction in the accumulation of de novo immunoglobulin gene somatic mutations upon vaccination. This reduction is associated with a significant decrease in the capacity of antibodies to target the viral glycoprotein, hemagglutinin (HA), and critical protective epitopes surrounding the HA receptor-binding domain. Immune escape by antigenic drift, in which viruses generate mutations in key antigenic epitopes, becomes highly exaggerated. Because of this reduced adaptability, most B cells activated in the elderly cohort target highly conserved but less potent epitopes. Given these findings, vaccines driving immunoglobulin gene somatic hypermutation should be a priority to protect elderly individuals.
Keywords: elderly population; immunoglobulin genes; influenza vaccine; monoclonal antibodies.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


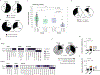

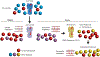
Comment in
-
Original Antigenic Sin: Friend or Foe in Developing a Broadly Cross-Reactive Vaccine to Influenza?Cell Host Microbe. 2019 Mar 13;25(3):354-355. doi: 10.1016/j.chom.2019.02.009. Cell Host Microbe. 2019. PMID: 30870620 Free PMC article.
References
-
- Babar MM, and Zaidi NU (2015). Protein sequence conservation and stable molecular evolution reveals influenza virus nucleoprotein as a universal druggable target. Infect Genet Evol 34, 200–210. - PubMed
MeSH terms
Substances
Grants and funding
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- UL1 TR002001/TR/NCATS NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- DP2 AI117921/AI/NIAID NIH HHS/United States
- U19 AI109946/AI/NIAID NIH HHS/United States
- T32 AI007090/AI/NIAID NIH HHS/United States
- R01 AI113047/AI/NIAID NIH HHS/United States
- HHSN272201400005C/AI/NIAID NIH HHS/United States
- P01 AI097092/AI/NIAID NIH HHS/United States
- P30 AG028716/AG/NIA NIH HHS/United States
- R01 AI117287/AI/NIAID NIH HHS/United States
- R01 AI128821/AI/NIAID NIH HHS/United States
- U19 AI082724/AI/NIAID NIH HHS/United States
- U19 AI057266/AI/NIAID NIH HHS/United States
- R01 AI108686/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

