Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila
- PMID: 30796039
- PMCID: PMC6442380
- DOI: 10.1101/gr.243832.118
Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila
Abstract
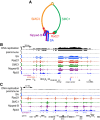
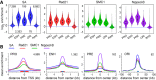
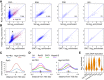
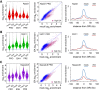
Cohesin consists of the SMC1-SMC3-Rad21 tripartite ring and the SA protein that interacts with Rad21. The Nipped-B protein loads cohesin topologically around chromosomes to mediate sister chromatid cohesion and facilitate long-range control of gene transcription. It is largely unknown how Nipped-B and cohesin associate specifically with gene promoters and transcriptional enhancers, or how sister chromatid cohesion is established. Here, we use genome-wide chromatin immunoprecipitation in Drosophila cells to show that SA and the Fs(1)h (BRD4) BET domain protein help recruit Nipped-B and cohesin to enhancers and DNA replication origins, whereas the MED30 subunit of the Mediator complex directs Nipped-B and Vtd in Drosophila (also known as Rad21) to promoters. All enhancers and their neighboring promoters are close to DNA replication origins and bind SA with proportional levels of cohesin subunits. Most promoters are far from origins and lack SA but bind Nipped-B and Rad21 with subproportional amounts of SMC1, indicating that they bind cohesin rings only part of the time. Genetic data show that Nipped-B and Rad21 function together with Fs(1)h to facilitate Drosophila development. These findings show that Nipped-B and cohesin are differentially targeted to enhancers and promoters, and suggest models for how SA and DNA replication help establish sister chromatid cohesion and facilitate enhancer-promoter communication. They indicate that SA is not an obligatory cohesin subunit but a factor that controls cohesin location on chromosomes.
© 2019 Pherson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Brca2, Pds5 and Wapl differentially control cohesin chromosome association and function.PLoS Genet. 2018 Feb 15;14(2):e1007225. doi: 10.1371/journal.pgen.1007225. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29447171 Free PMC article.
-
Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene.Mol Cell Biol. 2004 Apr;24(8):3100-11. doi: 10.1128/MCB.24.8.3100-3111.2004. Mol Cell Biol. 2004. PMID: 15060134 Free PMC article.
-
Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells.Chromosoma. 2008 Feb;117(1):51-66. doi: 10.1007/s00412-007-0125-5. Epub 2007 Oct 2. Chromosoma. 2008. PMID: 17909832 Free PMC article.
-
The Many Roles of Cohesin in Drosophila Gene Transcription.Trends Genet. 2019 Jul;35(7):542-551. doi: 10.1016/j.tig.2019.04.002. Epub 2019 May 23. Trends Genet. 2019. PMID: 31130395 Free PMC article. Review.
-
Cohesin, gene expression and development: lessons from Drosophila.Chromosome Res. 2009;17(2):185-200. doi: 10.1007/s10577-009-9022-5. Chromosome Res. 2009. PMID: 19308700 Free PMC article. Review.
Cited by
-
The Drosophila MLR COMPASS complex is essential for programming cis-regulatory information and maintaining epigenetic memory during development.Nucleic Acids Res. 2020 Apr 17;48(7):3476-3495. doi: 10.1093/nar/gkaa082. Nucleic Acids Res. 2020. PMID: 32052053 Free PMC article.
-
Paralogous synthetic lethality underlies genetic dependencies of the cancer-mutated gene STAG2.Life Sci Alliance. 2021 Aug 30;4(11):e202101083. doi: 10.26508/lsa.202101083. Print 2021 Nov. Life Sci Alliance. 2021. PMID: 34462321 Free PMC article.
-
SETting up the genome: KMT2D and KDM6A genomic function in the Kabuki syndrome craniofacial developmental disorder.Birth Defects Res. 2023 Dec 1;115(20):1885-1898. doi: 10.1002/bdr2.2253. Epub 2023 Oct 6. Birth Defects Res. 2023. PMID: 37800171 Free PMC article. Review.
-
Mechanisms of Enhancer-Promoter Interactions in Higher Eukaryotes.Int J Mol Sci. 2021 Jan 12;22(2):671. doi: 10.3390/ijms22020671. Int J Mol Sci. 2021. PMID: 33445415 Free PMC article. Review.
-
Insights into HP1a-Chromatin Interactions.Cells. 2020 Aug 9;9(8):1866. doi: 10.3390/cells9081866. Cells. 2020. PMID: 32784937 Free PMC article. Review.
References
-
- Brough R, Bajrami I, Vatcheva R, Natrajan R, Reis-Filho JS, Lord CJ, Ashworth A. 2012. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J 31: 1160–1176. 10.1038/emboj.2011.490 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
