Absence of Global Stress Regulation in Escherichia coli Promotes Pathoadaptation and Novel c-di-GMP-dependent Metabolic Capability
- PMID: 30796316
- PMCID: PMC6385356
- DOI: 10.1038/s41598-019-39580-w
Absence of Global Stress Regulation in Escherichia coli Promotes Pathoadaptation and Novel c-di-GMP-dependent Metabolic Capability
Abstract
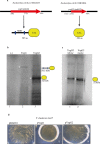
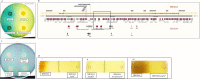
Pathoadaptive mutations linked to c-di-GMP signalling were investigated in neonatal meningitis-causing Escherichia coli (NMEC). The results indicated that NMEC strains deficient in RpoS (the global stress regulator) maintained remarkably low levels of c-di-GMP, a major bacterial sessility-motility switch. Deletion of ycgG2, shown here to encode a YcgG allozyme with c-di-GMP phosphodiesterase activity, and the restoration of RpoS led to a decrease in S-fimbriae, robustly produced in artificial urine, hinting that the urinary tract could serve as a habitat for NMEC. We showed that NMEC were skilled in aerobic citrate utilization in the presence of glucose, a property that normally does not exist in E. coli. Our data suggest that this metabolic novelty is a property of extraintestinal pathogenic E. coli since we reconstituted this ability in E. coli UTI89 (a cystitis isolate) via deactivation rpoS; additionally, a set of pyelonephritis E. coli isolates were shown here to aerobically use citrate in the presence of glucose. We found that the main reason for this metabolic capability is RpoS inactivation leading to the production of the citrate transporter CitT, exploited by NMEC for ferric citrate uptake dependent on YcgG2 (an allozyme with c-di-GMP phosphodiesterase activity).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Boone, D. R., Castenholz, R. W. & Garrity, G. M. Bergey’s manual of systematic bacteriology. 2nd edn, (Springer, 2001).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

