Within-Host Genomic Diversity of Candida albicans in Healthy Carriers
- PMID: 30796326
- PMCID: PMC6385308
- DOI: 10.1038/s41598-019-38768-4
Within-Host Genomic Diversity of Candida albicans in Healthy Carriers
Abstract
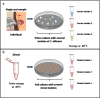
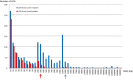
Genomic variations in Candida albicans, a major fungal pathogen of humans, have been observed upon exposure of this yeast to different stresses and experimental infections, possibly contributing to subsequent adaptation to these stress conditions. Yet, little is known about the extent of genomic diversity that is associated with commensalism, the predominant lifestyle of C. albicans in humans. In this study, we investigated the genetic diversity of C. albicans oral isolates recovered from healthy individuals, using multilocus sequencing typing (MLST) and whole genome sequencing. While MLST revealed occasional differences between isolates collected from a single individual, genome sequencing showed that they differed by numerous single nucleotide polymorphisms, mostly resulting from short-range loss-of-heterozygosity events. These differences were shown to have occurred upon human carriage of C. albicans rather than subsequent in vitro manipulation of the isolates. Thus, C. albicans intra-sample diversity appears common in healthy individuals, higher than that observed using MLST. We propose that diversifying lineages coexist in a single human individual, and this diversity can enable rapid adaptation under stress exposure. These results are crucial for the interpretation of longitudinal studies evaluating the evolution of the C. albicans genome.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract.J Clin Microbiol. 2006 May;44(5):1810-20. doi: 10.1128/JCM.44.5.1810-1820.2006. J Clin Microbiol. 2006. PMID: 16672411 Free PMC article.
-
Genetic diversity of Candida albicans isolates recovered from hospital environments and patients with severe acquired brain injuries.Infect Genet Evol. 2019 Dec;76:104068. doi: 10.1016/j.meegid.2019.104068. Epub 2019 Oct 12. Infect Genet Evol. 2019. PMID: 31614212
-
Genotyping Candida albicans from Candida leukoplakia and non-Candida leukoplakia shows no enrichment of multilocus sequence typing clades but enrichment of ABC genotype C in Candida leukoplakia.PLoS One. 2013 Sep 18;8(9):e73738. doi: 10.1371/journal.pone.0073738. eCollection 2013. PLoS One. 2013. PMID: 24058485 Free PMC article.
-
Molecular epidemiology, phylogeny and evolution of Candida albicans.Infect Genet Evol. 2014 Jan;21:166-78. doi: 10.1016/j.meegid.2013.11.008. Epub 2013 Nov 19. Infect Genet Evol. 2014. PMID: 24269341 Review.
-
Multilocus sequence typing of Candida albicans: strategies, data exchange and applications.Infect Genet Evol. 2004 Sep;4(3):243-52. doi: 10.1016/j.meegid.2004.06.002. Infect Genet Evol. 2004. PMID: 15450203 Review.
Cited by
-
EFG1, Everyone's Favorite Gene in Candida albicans: A Comprehensive Literature Review.Front Cell Infect Microbiol. 2022 Mar 22;12:855229. doi: 10.3389/fcimb.2022.855229. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35392604 Free PMC article. Review.
-
Immunosurveillance of Candida albicans commensalism by the adaptive immune system.Mucosal Immunol. 2022 May;15(5):829-836. doi: 10.1038/s41385-022-00536-5. Epub 2022 Jul 1. Mucosal Immunol. 2022. PMID: 35778599 Free PMC article. Review.
-
SNP-SVant: A Computational Workflow to Predict and Annotate Genomic Variants in Organisms Lacking Benchmarked Variants.Curr Protoc. 2024 May;4(5):e1046. doi: 10.1002/cpz1.1046. Curr Protoc. 2024. PMID: 38717471 Free PMC article.
-
Danish Whole-Genome-Sequenced Candida albicans and Candida glabrata Samples Fit into Globally Prevalent Clades.J Fungi (Basel). 2021 Nov 12;7(11):962. doi: 10.3390/jof7110962. J Fungi (Basel). 2021. PMID: 34829249 Free PMC article.
-
Molecular Diversity and Genetic Relatedness of Candida albicans Isolates from Birds in Hungary.Mycopathologia. 2021 May;186(2):237-244. doi: 10.1007/s11046-021-00527-3. Epub 2021 Jan 29. Mycopathologia. 2021. PMID: 33512664 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

