Circular RNAs are temporospatially regulated throughout development and ageing in the rat
- PMID: 30796328
- PMCID: PMC6385508
- DOI: 10.1038/s41598-019-38860-9
Circular RNAs are temporospatially regulated throughout development and ageing in the rat
Abstract
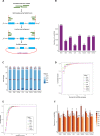
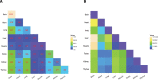
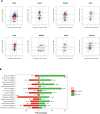
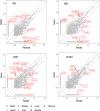
Circular RNAs (circRNAs) are covalently closed structural isoforms of linear mRNA which have been observed across a broad range of species and tissues. Here, we provide a comprehensive circRNAs expression catalogue for the rat including 8 organs of both sexes during 4 developmental stages using a public RNAseq dataset. These analyses revealed thousands of circular RNA species, many expressed in an organ-specific manner along with their host genes which were enriched with tissue-specific biological functions. A large number of circRNAs also displayed a developmental-dependent expression pattern and are accumulated during ageing. CircRNAs also displayed some sexually dimorphic expression, with gender associated differences observed in various tissues and developmental stages. These observations suggest that circRNAs are dynamically expressed in a spatial-, temporal- and gender-specific manner in mammals, and may have important biological function in differentiation, development and aging.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

