Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis
- PMID: 30796637
- PMCID: PMC6711945
- DOI: 10.1007/s13238-019-0610-7
Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis
Abstract
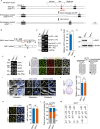
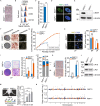
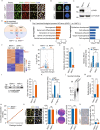
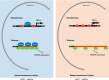
RAP1 is a well-known telomere-binding protein, but its functions in human stem cells have remained unclear. Here we generated RAP1-deficient human embryonic stem cells (hESCs) by using CRISPR/Cas9 technique and obtained RAP1-deficient human mesenchymal stem cells (hMSCs) and neural stem cells (hNSCs) via directed differentiation. In both hMSCs and hNSCs, RAP1 not only negatively regulated telomere length but also acted as a transcriptional regulator of RELN by tuning the methylation status of its gene promoter. RAP1 deficiency enhanced self-renewal and delayed senescence in hMSCs, but not in hNSCs, suggesting complicated lineage-specific effects of RAP1 in adult stem cells. Altogether, these results demonstrate for the first time that RAP1 plays both telomeric and nontelomeric roles in regulating human stem cell homeostasis.
Keywords: RAP1; RELN; methylation; stem cell; telomere.
Conflict of interest statement
Xing Zhang, Zunpeng Liu, Xiaoqian Liu, Si Wang, Yiyuan Zhang, Xiaojuan He, Shuhui Sun, Shuai Ma, Ng Shyh-Chang, Feng Liu, Qiang Wang, Xiaoqun Wang, Lin Liu, Weiqi Zhang, Moshi Song, Guang-Hui Liu and Jing Qu declare that they have no conflict of interest. All institutional and national guidelines for the care and use of laboratory animals were followed.
Figures



Similar articles
-
Inhibition of the histone demethylase Kdm5b promotes neurogenesis and derepresses Reln (reelin) in neural stem cells from the adult subventricular zone of mice.Mol Biol Cell. 2016 Feb 15;27(4):627-39. doi: 10.1091/mbc.E15-07-0513. Epub 2016 Jan 6. Mol Biol Cell. 2016. PMID: 26739753 Free PMC article.
-
Decoding telomere protein Rap1: Its telomeric and nontelomeric functions and potential implications in diabetic cardiomyopathy.Cell Cycle. 2017 Oct 2;16(19):1765-1773. doi: 10.1080/15384101.2017.1371886. Epub 2017 Aug 30. Cell Cycle. 2017. PMID: 28853973 Free PMC article. Review.
-
Roles of Reelin/Disabled1 pathway on functional recovery of hemiplegic mice after neural cell transplantation; Reelin promotes migration toward motor cortex and maturation to motoneurons of neural grafts.Exp Neurol. 2019 Oct;320:112970. doi: 10.1016/j.expneurol.2019.112970. Epub 2019 Jun 8. Exp Neurol. 2019. PMID: 31185198
-
Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity.Cell Rep. 2013 Jun 27;3(6):1847-56. doi: 10.1016/j.celrep.2013.05.032. Epub 2013 Jun 20. Cell Rep. 2013. PMID: 23791522 Free PMC article.
-
Role of Rap1 in DNA damage response: implications in stem cell homeostasis and cancer.Exp Hematol. 2020 Oct;90:12-17. doi: 10.1016/j.exphem.2020.08.009. Epub 2020 Aug 26. Exp Hematol. 2020. PMID: 32858091 Review.
Cited by
-
Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction.Protein Cell. 2020 Jan;11(1):1-22. doi: 10.1007/s13238-019-0623-2. Epub 2019 Apr 30. Protein Cell. 2020. PMID: 31037510 Free PMC article.
-
Roles of chromatin and genome instability in cellular senescence and their relevance to ageing and related diseases.Nat Rev Mol Cell Biol. 2024 Dec;25(12):979-1000. doi: 10.1038/s41580-024-00775-3. Epub 2024 Oct 3. Nat Rev Mol Cell Biol. 2024. PMID: 39363000 Review.
-
A human circulating immune cell landscape in aging and COVID-19.Protein Cell. 2020 Oct;11(10):740-770. doi: 10.1007/s13238-020-00762-2. Epub 2020 Aug 11. Protein Cell. 2020. PMID: 32780218 Free PMC article.
-
SIRT3 consolidates heterochromatin and counteracts senescence.Nucleic Acids Res. 2021 May 7;49(8):4203-4219. doi: 10.1093/nar/gkab161. Nucleic Acids Res. 2021. PMID: 33706382 Free PMC article.
-
Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions.mSphere. 2020 Feb 26;5(1):e00027-20. doi: 10.1128/mSphere.00027-20. mSphere. 2020. PMID: 32102938 Free PMC article.
References
-
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat Struct Mol Biol. 2012;19:948–956. - PubMed
-
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. - PubMed
-
- Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350:1193–1198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

