Polypyrimidine Tract-Binding Protein Regulates Enterovirus 71 Translation Through Interaction with the Internal Ribosomal Entry Site
- PMID: 30796736
- PMCID: PMC6420457
- DOI: 10.1007/s12250-019-00089-1
Polypyrimidine Tract-Binding Protein Regulates Enterovirus 71 Translation Through Interaction with the Internal Ribosomal Entry Site
Abstract
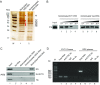
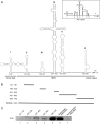
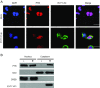
Enterovirus 71 (EV71), a major causative agent of hand, foot, and mouth disease, has caused periodic infection outbreaks in children in the Asia-Pacific region. In order to describe the largely unknown life cycle of EV71, the molecular basis of its virus-host interactions must first be determined. The 5' untranslated region of EV71 contains a cloverleaf-like structure and internal ribosomal entry site (IRES), which play an important role in transcription and translation of viral protein. We found that polypyrimidine tract-binding protein 1 (PTB) bound to the IRES of EV71. RNA recognition motifs 1 and 2 of PTB were responsible for its binding to the EV71 IRES. Moreover, PTB protein was shuttled from nucleus to cytoplasm after EV71 infection. Additionally, IRES activity and viral protein production were inhibited by PTB knockdown. These results suggest that PTB interacts with the EV71 IRES, and positively regulates viral protein translation.
Keywords: EV71; IRES; PTB; Translation.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures


Similar articles
-
Nuclear Protein Sam68 Interacts with the Enterovirus 71 Internal Ribosome Entry Site and Positively Regulates Viral Protein Translation.J Virol. 2015 Oct;89(19):10031-43. doi: 10.1128/JVI.01677-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202240 Free PMC article.
-
SIRT1 inhibits EV71 genome replication and RNA translation by interfering with the viral polymerase and 5'UTR RNA.J Cell Sci. 2016 Dec 15;129(24):4534-4547. doi: 10.1242/jcs.193698. Epub 2016 Nov 14. J Cell Sci. 2016. PMID: 27875274 Free PMC article.
-
Enterovirus 71 Activates GADD34 via Precursor 3CD to Promote IRES-Mediated Viral Translation.Microbiol Spectr. 2022 Feb 23;10(1):e0138821. doi: 10.1128/spectrum.01388-21. Epub 2022 Jan 5. Microbiol Spectr. 2022. PMID: 34985336 Free PMC article.
-
Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein.Biochem Soc Trans. 2008 Aug;36(Pt 4):641-7. doi: 10.1042/BST0360641. Biochem Soc Trans. 2008. PMID: 18631133 Review.
-
Activation of Host Cellular Signaling and Mechanism of Enterovirus 71 Viral Proteins Associated with Hand, Foot and Mouth Disease.Viruses. 2022 Oct 4;14(10):2190. doi: 10.3390/v14102190. Viruses. 2022. PMID: 36298746 Free PMC article. Review.
Cited by
-
Translation control of Enterovirus A71 gene expression.J Biomed Sci. 2020 Jan 8;27(1):22. doi: 10.1186/s12929-019-0607-9. J Biomed Sci. 2020. PMID: 31910851 Free PMC article. Review.
-
Host neuronal PRSS3 interacts with enterovirus A71 3A protein and its role in viral replication.Sci Rep. 2022 Jul 27;12(1):12846. doi: 10.1038/s41598-022-17272-2. Sci Rep. 2022. PMID: 35896602 Free PMC article.
-
Parechovirus: neglected for too long?J Virol. 2025 Apr 15;99(4):e0184624. doi: 10.1128/jvi.01846-24. Epub 2025 Mar 25. J Virol. 2025. PMID: 40130875 Free PMC article. Review.
-
Heat Shock Protein A6, a Novel HSP70, Is Induced During Enterovirus A71 Infection to Facilitate Internal Ribosomal Entry Site-Mediated Translation.Front Microbiol. 2021 May 7;12:664955. doi: 10.3389/fmicb.2021.664955. eCollection 2021. Front Microbiol. 2021. PMID: 34025620 Free PMC article.
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

