Light-sheet fluorescence expansion microscopy: fast mapping of neural circuits at super resolution
- PMID: 30796881
- PMCID: PMC6368534
- DOI: 10.1117/1.NPh.6.1.015005
Light-sheet fluorescence expansion microscopy: fast mapping of neural circuits at super resolution
Abstract
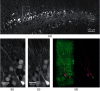
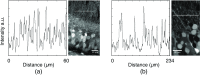
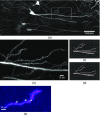
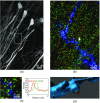
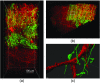
The goal of understanding the architecture of neural circuits at the synapse level with a brain-wide perspective has powered the interest in high-speed and large field-of-view volumetric imaging at subcellular resolution. Here, we developed a method combining tissue expansion and light-sheet fluorescence microscopy to allow extended volumetric super resolution high-speed imaging of large mouse brain samples. We demonstrate the capabilities of this method by performing two color fast volumetric super resolution imaging of mouse CA1 and dentate gyrus molecular-, granule cell-, and polymorphic layers. Our method enables an exact evaluation of granule cell and neurite morphology within the context of large cell ensembles spanning several orders of magnitude in resolution. We found that imaging a brain region of in super resolution using light-sheet fluorescence expansion microscopy is about 17-fold faster than imaging the same region by a current state-of-the-art high-resolution confocal laser scanning microscope.
Keywords: connectomics; dentate gyrus; light-sheet fluorescence microscopy; super resolution; tissue expansion.
Figures

References
-
- Ramón S., Cajal Y., Textura del sistema nervioso del hombre y de los vertebrados, Imprenta y Librería de Nicolás Moya, Madrid: (1899–1904).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

