Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation
- PMID: 30797686
- PMCID: PMC6544384
- DOI: 10.1016/j.molcel.2019.01.014
Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation
Abstract
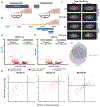
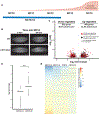
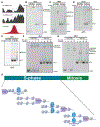
Reprogramming cell fate during the first stages of embryogenesis requires that transcriptional activators gain access to the genome and remodel the zygotic transcriptome. Nonetheless, it is not clear whether the continued activity of these pioneering factors is required throughout zygotic genome activation or whether they are only required early to establish cis-regulatory regions. To address this question, we developed an optogenetic strategy to rapidly and reversibly inactivate the master regulator of genome activation in Drosophila, Zelda. Using this strategy, we demonstrate that continued Zelda activity is required throughout genome activation. We show that Zelda binds DNA in the context of nucleosomes and suggest that this allows Zelda to occupy the genome despite the rapid division cycles in the early embryo. These data identify a powerful strategy to inactivate transcription factor function during development and suggest that reprogramming in the embryo may require specific, continuous pioneering functions to activate the genome.
Keywords: Drosophila; MZT; ZGA; Zelda; maternal-to-zygotic transition; optogenetic; pioneer factor; transcription; zygotic genome activation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures

Similar articles
-
GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo.Elife. 2021 Mar 15;10:e66668. doi: 10.7554/eLife.66668. Elife. 2021. PMID: 33720012 Free PMC article.
-
Premature translation of the Drosophila zygotic genome activator Zelda is not sufficient to precociously activate gene expression.G3 (Bethesda). 2022 Aug 25;12(9):jkac159. doi: 10.1093/g3journal/jkac159. G3 (Bethesda). 2022. PMID: 35876878 Free PMC article.
-
CLAMP regulates zygotic genome activation in Drosophila embryos.Genetics. 2021 Oct 2;219(2):iyab107. doi: 10.1093/genetics/iyab107. Genetics. 2021. PMID: 34849887 Free PMC article.
-
Regulatory principles governing the maternal-to-zygotic transition: insights from Drosophila melanogaster.Open Biol. 2018 Dec;8(12):180183. doi: 10.1098/rsob.180183. Open Biol. 2018. PMID: 30977698 Free PMC article. Review.
-
Regulation and Function of Maternal Gene Products During the Maternal-to-Zygotic Transition in Drosophila.Curr Top Dev Biol. 2015;113:43-84. doi: 10.1016/bs.ctdb.2015.06.007. Epub 2015 Aug 14. Curr Top Dev Biol. 2015. PMID: 26358870 Review.
Cited by
-
Minimal synthetic enhancers reveal control of the probability of transcriptional engagement and its timing by a morphogen gradient.Cell Syst. 2023 Mar 15;14(3):220-236.e3. doi: 10.1016/j.cels.2022.12.008. Epub 2023 Jan 24. Cell Syst. 2023. PMID: 36696901 Free PMC article.
-
The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development.Cells. 2019 Jul 29;8(8):788. doi: 10.3390/cells8080788. Cells. 2019. PMID: 31362461 Free PMC article. Review.
-
Histone variant H2A.Z regulates zygotic genome activation.Nat Commun. 2021 Dec 1;12(1):7002. doi: 10.1038/s41467-021-27125-7. Nat Commun. 2021. PMID: 34853314 Free PMC article.
-
GAGA factor: a multifunctional pioneering chromatin protein.Cell Mol Life Sci. 2021 May;78(9):4125-4141. doi: 10.1007/s00018-021-03776-z. Epub 2021 Feb 2. Cell Mol Life Sci. 2021. PMID: 33528710 Free PMC article. Review.
-
An Epigenetic Priming Mechanism Mediated by Nutrient Sensing Regulates Transcriptional Output during C. elegans Development.Curr Biol. 2021 Feb 22;31(4):809-826.e6. doi: 10.1016/j.cub.2020.11.060. Epub 2020 Dec 22. Curr Biol. 2021. PMID: 33357451 Free PMC article.
References
-
- Anderson KV, and Lengyel JA (1979). Rates of synthesis of major classes of RNA in Drosophila embryos. Dev Biol 70, 217–231. - PubMed
-
- ten Bosch JR, Benavides JA, and Cline TW (2006). The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967–1977. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

