MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility
- PMID: 30797712
- PMCID: PMC6444028
- DOI: 10.1016/j.ebiom.2018.12.049
MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility
Abstract
Background: The low expression of miR93/25 (members of miR-106b~25 cluster) promoted the invasion and metastasis of colon cancer cells, which predicted poor survival. However, the role of miR-106b-5p, the member of miR-106b~25 cluster, in colorectal cancer (CRC) remains unclear.
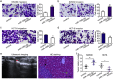
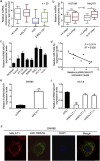
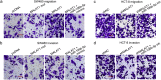
Methods: Bioinformatics methods were used to predict the potential pairs of lncRNA-miRNA-mRNA. In situ hybridization and qPCR were used to evaluate the expression of MALAT1 and miR-106b-5p in the paraffin-embedded normal and CRC tissues. Kaplan-Meier analysis with the log-rank test was used for survival analyses. Immunohistochemistry staining was applied to investigate the expression of SLAIN2. Fluorescence recovery after photobleaching assay was applied to observe the microtubule (MT) mobility. In vitro and in vivo invasion and metastasis assays were used to explore the function of MALAT1/miR-106b-5p/SLAIN2 in the progression of CRC.
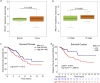
Findings: miR-106b-5p was identified as a suppressor in CRC. Functionally, ectopic or silencing the expression of miR-106b-5p inhibited or promoted the invasion and metastasis of CRC cells in vitro and in vivo. The long non-coding RNA MALAT1 regulated the miR-106b-5p expression and further mediated the mobility of SLAIN2-related MTs by functioning as a competing endogenous RNA in vitro and in vivo, which resulted in the progression of CRC. Clinically, low miR-106b-5p expression predicted poor survival of CRC patients, especially in combination with high MALAT1/ SLAIN2 expression.
Interpretation: miR-106b-5p served as a suppressor in combination with MALAT1/miR-106b-5p/SLAIN2, which might be a group of potential prognostic biomarkers in the prognosis of CRC. FUND: This work was supported by National Program Project for Precision Medicine in National Research and Development Plan of China (2016YFC0905300), National Natural Science Foundation of China (81572930), National Key Research and Development Program of the Ministry of Science and Technology of China (2016YFC0905303, 2016YFC1303200), Beijing Science and Technology Program (D17110002617004), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT32012), CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001), Incentive Fund for Academic Leaders of Oncology Hospital, Chinese Academy of Medical Sciences (RC2016003), and Beijing Hope Run Special Fund from Cancer Foundation of China (LC2017A19). The project of Shanghai Jiaotong Univversity (YG2017QN30).
Keywords: Colorectal cancer progression; MALAT1; Microtubules mobility; miR-106ba-5p.
Copyright © 2018. Published by Elsevier B.V.
Figures








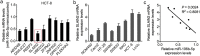
References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer Statistics. 2018;68(1):7–30. - PubMed
-
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Li Q., Zou C., Zou C. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335(1):168–174. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

