Assessment and Maintenance of Unigametic Germline Inheritance for C. elegans
- PMID: 30799227
- PMCID: PMC6435406
- DOI: 10.1016/j.devcel.2019.01.020
Assessment and Maintenance of Unigametic Germline Inheritance for C. elegans
Abstract
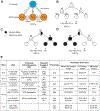
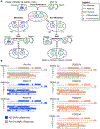
The recent work of Besseling and Bringmann (2016) identified a molecular intervention for C. elegans in which premature segregation of maternal and paternal chromosomes in the fertilized oocyte can produce viable animals exhibiting a non-Mendelian inheritance pattern. Overexpression in embryos of a single protein regulating chromosome segregation (GPR-1) provides a germline derived clonally from a single parental gamete. We present a collection of strains and cytological assays to consistently generate and track non-Mendelian inheritance. These tools allow reproducible and high-frequency (>80%) production of non-Mendelian inheritance, the facile and simultaneous homozygosis for all nuclear chromosomes in a single generation, the precise exchange of nuclear and mitochondrial genomes between strains, and the assessments of non-canonical mitosis events. We show the utility of these strains by demonstrating a rapid assessment of cell lineage requirements (AB versus P1) for a set of genes (lin-2, lin-3, lin-12, and lin-31) with roles in C. elegans vulval development.
Keywords: C. elegans; genetic engineering; inheritance; mitosis; mosaic; non-Mendelian; synthetic biology.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures



Comment in
-
Still Searching for Specialized Ribosomes.Dev Cell. 2019 Mar 25;48(6):744-746. doi: 10.1016/j.devcel.2019.03.005. Dev Cell. 2019. PMID: 30913404
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

