Prophage Excision in Streptococcus pneumoniae Serotype 19A ST320 Promote Colonization: Insight Into Its Evolution From the Ancestral Clone Taiwan 19F-14 (ST236)
- PMID: 30800118
- PMCID: PMC6375853
- DOI: 10.3389/fmicb.2019.00205
Prophage Excision in Streptococcus pneumoniae Serotype 19A ST320 Promote Colonization: Insight Into Its Evolution From the Ancestral Clone Taiwan 19F-14 (ST236)
Abstract
Streptococcus pneumoniae 19A ST320, a multidrug-resistant strain with high disease severity that notoriously spread before the use of expanded pneumococcal conjugate vaccines, was derived from a capsular switching event between an international strain Taiwan 19F-14 (ST236) and a serotype 19A strain. However, the molecular mechanisms underlying the adaptive evolution of 19F ST236 to 19A ST320 are unknown. In this study, we compared 19A ST320 to its ancestral clone, 19F ST236, in terms of adherence to respiratory epithelial cells, whole transcriptome, and ability to colonize a young mouse model. Serotype 19A ST320 showed five-fold higher adherence to A549 cells than serotype 19F ST236. High-throughput mRNA sequencing identified a prophage region located between dnaN and ychF in both strains; however, the genes in this region were expressed at significantly higher levels in 19A ST320 than in 19F ST236. Analysis by polymerase chain reaction (PCR) showed that the prophage is able to spontaneously excise from the chromosome and form a circular episome in 19A ST320, but not in 19F ST236. Deletion of the integrase in the prophage of 19A ST320 decreased spontaneous excision and cell adherence, which were restored by complementation. Competition experiments in mice showed that the integrase mutant was six-fold less competitive than the 19A ST320 parent (competitive index [CI]: 0.16; p = 0.02). The 19A ST320 prophage-deleted strain did not change cell adherence capacity, whereas prophage integration strains (integrase mutant and 19F) had decreased expression of the down-stream ychF gene compared to that of 19A ST320. Further deletion of ychF significantly reduced cell adherence. In conclusions, these findings suggest that spontaneous prophage induction confers a competitive advantage to virulent pneumococci.
Keywords: Streptococcus pneumoniae; adherence; integrase; mRNA sequencing; phage.
Figures
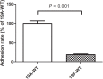



 , left Y-axis) and viable CFUs counts on blood agar (
, left Y-axis) and viable CFUs counts on blood agar ( , right Y-axis) of serotype 19A ST320 wild type (19A-WT) and its integrase deletion mutant (19A-Δint). (B) Adherence of serotype 19A ST320 wild type (19A-WT), the integrase mutant (19A-Δint), and the complementation strain (19A-Δint::int) (wild type vs. mutant, p < 0.002, n = 4; wild type vs. complementation, p = 0.8, n = 4). (C) Intranasal challenge of 3-week-old female BALB/c mice with equal inocula of the bacterial strains. Each symbol represents the competitive index (CI) for an individual animal. A competitive index (CI) was calculated based on the ratio of the competing bacterial strains recovered by nasal lavage, normalized to the ratio of the respective bacteria in the inoculum = (outputtest strain/outputwild type)/(inputtest strain/inputwild type). The integrase mutant showed a lower CI compared to serotype 19A ST320 (p = 0.02, n = 4).
, right Y-axis) of serotype 19A ST320 wild type (19A-WT) and its integrase deletion mutant (19A-Δint). (B) Adherence of serotype 19A ST320 wild type (19A-WT), the integrase mutant (19A-Δint), and the complementation strain (19A-Δint::int) (wild type vs. mutant, p < 0.002, n = 4; wild type vs. complementation, p = 0.8, n = 4). (C) Intranasal challenge of 3-week-old female BALB/c mice with equal inocula of the bacterial strains. Each symbol represents the competitive index (CI) for an individual animal. A competitive index (CI) was calculated based on the ratio of the competing bacterial strains recovered by nasal lavage, normalized to the ratio of the respective bacteria in the inoculum = (outputtest strain/outputwild type)/(inputtest strain/inputwild type). The integrase mutant showed a lower CI compared to serotype 19A ST320 (p = 0.02, n = 4).
Similar articles
-
Expansion and evolution of Streptococcus pneumoniae serotype 19A ST320 clone as compared to its ancestral clone, Taiwan19F-14 (ST236).J Infect Dis. 2013 Jul 15;208(2):203-10. doi: 10.1093/infdis/jit145. Epub 2013 Apr 4. J Infect Dis. 2013. PMID: 23559465
-
Acute otitis media caused by Streptococcus pneumoniae serotype 19A ST320 clone: epidemiological and clinical characteristics.J Microbiol Immunol Infect. 2018 Jun;51(3):337-343. doi: 10.1016/j.jmii.2016.08.002. Epub 2016 Dec 19. J Microbiol Immunol Infect. 2018. PMID: 28087317
-
Phylogenomic insights into evolutionary trajectories of multidrug resistant S. pneumoniae CC271 over a period of 14 years in China.Genome Med. 2023 Jul 4;15(1):46. doi: 10.1186/s13073-023-01200-8. Genome Med. 2023. PMID: 37403170 Free PMC article.
-
The impact of 10-valent and 13-valent pneumococcal conjugate vaccines on serotype 19A invasive pneumococcal disease.Expert Rev Vaccines. 2015;14(10):1359-66. doi: 10.1586/14760584.2015.1075884. Epub 2015 Aug 7. Expert Rev Vaccines. 2015. PMID: 26289973 Review.
-
Pneumococcal disease caused by serotype 19A: review of the literature and implications for future vaccine development.Vaccine. 2010 Jun 11;28(26):4249-59. doi: 10.1016/j.vaccine.2010.04.020. Epub 2010 Apr 21. Vaccine. 2010. PMID: 20416266 Review.
Cited by
-
Infection with Bacteroides Phage BV01 Alters the Host Transcriptome and Bile Acid Metabolism in a Common Human Gut Microbe.Cell Rep. 2020 Sep 15;32(11):108142. doi: 10.1016/j.celrep.2020.108142. Cell Rep. 2020. PMID: 32937127 Free PMC article.
-
Serotype Distribution, Antimicrobial Susceptibility, Multilocus Sequencing Type and Virulence of Invasive Streptococcus pneumoniae in China: A Six-Year Multicenter Study.Front Microbiol. 2022 Jan 13;12:798750. doi: 10.3389/fmicb.2021.798750. eCollection 2021. Front Microbiol. 2022. PMID: 35095809 Free PMC article.
-
Streptococcus pneumoniae: a Plethora of Temperate Bacteriophages With a Role in Host Genome Rearrangement.Front Cell Infect Microbiol. 2021 Nov 18;11:775402. doi: 10.3389/fcimb.2021.775402. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34869076 Free PMC article.
-
Bacterial factors required for Streptococcus pneumoniae coinfection with influenza A virus.J Biomed Sci. 2021 Aug 27;28(1):60. doi: 10.1186/s12929-021-00756-0. J Biomed Sci. 2021. PMID: 34452635 Free PMC article.
-
Pan-Genome-Wide Association Study of Serotype 19A Pneumococci Identifies Disease-Associated Genes.Microbiol Spectr. 2023 Aug 17;11(4):e0407322. doi: 10.1128/spectrum.04073-22. Epub 2023 Jun 26. Microbiol Spectr. 2023. PMID: 37358412 Free PMC article.
References
LinkOut - more resources
Full Text Sources

