Cell Type Diversity in Hepatitis B Virus RNA Splicing and Its Regulation
- PMID: 30800119
- PMCID: PMC6375855
- DOI: 10.3389/fmicb.2019.00207
Cell Type Diversity in Hepatitis B Virus RNA Splicing and Its Regulation
Abstract
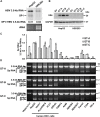
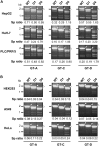
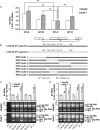
Although RNA splicing of hepatitis B virus (HBV) is a commonly observed in livers of hepatitis B patients as well as in the cultured cells replicating the viral genome, its biological significance in the HBV life cycle and the detailed regulatory mechanisms are still largely unclear. In this study, we found cell-type dependency of HBV splicing of the 3.5 kb pregenomic RNA, which is efficiently spliced in human hepatoma cells but not in cells derived from human hepatic stellate, mouse hepatoma and human non-hepatic cells. It may be likely that RNA splicing is one of the determinants of host range restriction of HBV. Given the finding indicating the difference in cell-type dependency of the splicing efficiency between HBV and simian virus 40, we carried out intron-swapping experiments. The results suggest the presence of putative exonic splicing enhancer that possibly works in the cell-type dependent fashion. Together with further mutational analyses, a novel 50-nt intronic splicing silencer, whose secondary structure is well conserved among the HBV strains, was identified. It appears that this intronic silencer functions effectively independent of cell backgrounds.
Keywords: alternative splicing; cis-acting element; exonic enhancer; hepatitis B virus; intronic silencer.
Figures


References
-
- Candotti D., Allain J.-P. (2017). Biological and clinical significance of hepatitis B virus RNA splicing: an update. Ann. Blood 2 1–14.
-
- Chida T., Ito M., Nakashima K., Kanegae Y., Aoshima T., Takabayashi S., et al. (2017). Critical role of CREBH-mediated induction of transforming growth factor beta2 by hepatitis C virus infection in fibrogenic responses in hepatic stellate cells. Hepatology 66 1430–1443. 10.1002/hep.29319 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

