Identification of Potential Lead Molecules for Zika Envelope Protein from In Silico Perspective
- PMID: 30800249
- PMCID: PMC6359704
Identification of Potential Lead Molecules for Zika Envelope Protein from In Silico Perspective
Abstract
Background: Zika virus is the family member of flavivirus with no reported clinically approved drugs or vaccines in the market till date. This virus is spread by Aedes mosquitoes, and can also be transmitted through sexual contact or blood transfusions. There are reported medical conditions like microcephaly among new-borns delivered by infected pregnant women. The envelope protein of Zika virus is associated with virulence, tropism, mediation of receptor binding and membrane fusion. ED1-EDII domain (K1 loop pocket) is an integral part of the envelope protein and a potential drug target. In the present study, the purpose was to identify the potential lead molecules to dock against K1 loop which could be later considered as flavivirus entry inhibitors.
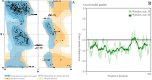
Methods: Multiple sequence alignment method was considered for the analysis of indels in envelope protein. Phylogenetic tree was constructed based on the alignment. Aliphatic index, GRAVY scores and hydropathy plot of the envelope proteins were calculated for the flavivirus family members. Zika envelope protein was homology modeled and considered for protein-ligand docking analysis with chemical compounds of known functions.
Results: As per in silico based analysis, the envelope protein of Zika virus is highly hydrophilic with the least number of amino acid deletions compared to rest of the family members. During docking studies, it was observed that compounds like NITD, compound 6, P02, Doxytetracycline and Rolitetracycline show better binding affinity with Zika envelope protein compared to dengue virus.
Conclusion: These better binding compounds could be the promising lead molecules for Zika envelope protein which could better block the viral entry.
Keywords: Aedes; Dengue virus; Envelope protein; Flavivirus; Microcephaly; Zika virus.
Conflict of interest statement
Conflict of Interest The authors declare they have no conflicts of interest.
Figures






References
-
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 1952;46(5):509–520. - PubMed
-
- Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg 1952;46(5):521–534. - PubMed
-
- Azari-Hamidian S. Checklist of Iranian mosquitoes (Diptera: Culicidae). J Vector Ecol 2007;32(2):235–242. - PubMed
-
- Mardani M. Update on Zika virus infections. Arch Clin Infect Dis 2016;11(2):69–71.
LinkOut - more resources
Full Text Sources
Research Materials
