Salinity Is a Key Determinant for Soil Microbial Communities in a Desert Ecosystem
- PMID: 30801023
- PMCID: PMC6372838
- DOI: 10.1128/mSystems.00225-18
Salinity Is a Key Determinant for Soil Microbial Communities in a Desert Ecosystem
Abstract
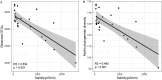
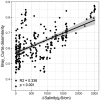
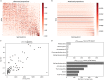
Soil salinization is a growing environmental problem caused by both natural and human activities. Excessive salinity in soil suppresses growth, decreases species diversity, and alters the community composition of plants; however, the effect of salinity on soil microbial communities is poorly understood. Here, we characterize the soil microbial community along a natural salinity gradient in Gurbantunggut Desert, Northwestern China. Microbial diversity linearly decreased with increases in salinity, and community dissimilarity significantly increased with salinity differences. Soil salinity showed a strong effect on microbial community dissimilarity, even after controlling for the effects of spatial distance and other environmental variables. Microbial phylotypes (n = 270) belonging to Halobacteria, Nitriliruptoria, [Rhodothermi], Gammaproteobacteria, and Alphaproteobacteria showed a high-salinity niche preference. Out of nine potential phenotypes predicted by BugBase, oxygen-related phenotypes showed a significant relationship with salinity content. To explore the community assembly processes, we used null models of within-community (nearest-taxon index [NTI]) and between-community (βNTI) phylogenetic composition. NTI showed a significantly negative relationship with salinity, suggesting that the microbial community was less phylogenetically clustered in more-saline soils. βNTI, the between-community analogue of NTI, showed that deterministic processes have overtaken stochastic processes across all sites, suggesting the importance of environmental filtering in microbial community assembly. Taken together, these results suggest the importance of salinity in soil microbial community composition and assembly processes in a desert ecosystem. IMPORTANCE Belowground microorganisms are indispensable components for nutrient cycling in desert ecosystems, and understanding how they respond to increased salinity is essential for managing and ameliorating salinization. Our sequence-based data revealed that microbial diversity decreased with increasing salinity, and certain salt-tolerant phylotypes and phenotypes showed a positive relationship with salinity. Using a null modeling approach to estimate microbial community assembly processes along a salinity gradient, we found that salinity imposed a strong selection pressure on the microbial community, which resulted in a dominance of deterministic processes. Studying microbial diversity and community assembly processes along salinity gradients is essential in understanding the fundamental ecological processes in desert ecosystems affected by salinization.
Keywords: community assembly processes; community diversity; desert ecosystem; microbial phenotypes; salinity.
Figures


Similar articles
-
Depth-dependent responses of soil bacterial communities to salinity in an arid region.Sci Total Environ. 2024 Nov 1;949:175129. doi: 10.1016/j.scitotenv.2024.175129. Epub 2024 Jul 30. Sci Total Environ. 2024. PMID: 39084388
-
The influence of soil salinization, induced by the backwater effect of the Yellow River, on microbial community dynamics and ecosystem functioning in arid regions.Environ Res. 2024 Dec 1;262(Pt 1):119854. doi: 10.1016/j.envres.2024.119854. Epub 2024 Aug 27. Environ Res. 2024. PMID: 39197488
-
Spatiotemporal Dynamics of Bacterial Community Assembly and Co-Occurrence Patterns in Biological Soil Crusts of Desert Ecosystems.Microorganisms. 2025 Feb 18;13(2):446. doi: 10.3390/microorganisms13020446. Microorganisms. 2025. PMID: 40005811 Free PMC article.
-
Microbial diversity and functions in saline soils: A review from a biogeochemical perspective.J Adv Res. 2024 May;59:129-140. doi: 10.1016/j.jare.2023.06.015. Epub 2023 Jun 29. J Adv Res. 2024. PMID: 37392974 Free PMC article. Review.
-
Microbiomics of Namib Desert habitats.Extremophiles. 2020 Jan;24(1):17-29. doi: 10.1007/s00792-019-01122-7. Epub 2019 Aug 2. Extremophiles. 2020. PMID: 31376000 Review.
Cited by
-
Planting Cyperus esculentus augments soil microbial biomass and diversity, but not enzymatic activities.PeerJ. 2022 Oct 13;10:e14199. doi: 10.7717/peerj.14199. eCollection 2022. PeerJ. 2022. PMID: 36258793 Free PMC article.
-
Metagenomic Insights into Microbial Community Structure, Function, and Salt Adaptation in Saline Soils of Arid Land, China.Microorganisms. 2022 Nov 3;10(11):2183. doi: 10.3390/microorganisms10112183. Microorganisms. 2022. PMID: 36363774 Free PMC article.
-
Fungal diversities and community assembly processes show different biogeographical patterns in forest and grassland soil ecosystems.Front Microbiol. 2023 Feb 1;14:1036905. doi: 10.3389/fmicb.2023.1036905. eCollection 2023. Front Microbiol. 2023. PMID: 36819045 Free PMC article.
-
Different living environments drive deterministic microbial community assemblages in the gut of Alpine musk deer (Moschus chrysogaster).Front Microbiol. 2023 Jan 13;13:1108405. doi: 10.3389/fmicb.2022.1108405. eCollection 2022. Front Microbiol. 2023. PMID: 36713154 Free PMC article.
-
Different responses of abundant and rare bacterial composition to groundwater depth and reduced nitrogen application in summer maize field.Front Microbiol. 2023 Oct 13;14:1220731. doi: 10.3389/fmicb.2023.1220731. eCollection 2023. Front Microbiol. 2023. PMID: 37901810 Free PMC article.
References
-
- Pan C, Liu C, Zhao H, Wang Y. 2013. Changes of soil physico-chemical properties and enzyme activities in relation to grassland salinization. Eur J Soil Biol 55:13–19. doi:10.1016/j.ejsobi.2012.09.009. - DOI
-
- Bui E. 2013. Soil salinity: a neglected factor in plant ecology and biogeography. J Arid Environ 92:14–25. doi:10.1016/j.jaridenv.2012.12.014. - DOI
-
- Rath KM, Rousk J. 2015. Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: a review. Soil Biol Biochem 81:108–123. doi:10.1016/j.soilbio.2014.11.001. - DOI
LinkOut - more resources
Full Text Sources
