Orphan Genes Shared by Pathogenic Genomes Are More Associated with Bacterial Pathogenicity
- PMID: 30801025
- PMCID: PMC6372840
- DOI: 10.1128/mSystems.00290-18
Orphan Genes Shared by Pathogenic Genomes Are More Associated with Bacterial Pathogenicity
Abstract
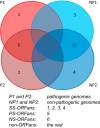
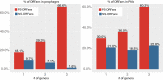
Orphan genes (also known as ORFans [i.e., orphan open reading frames]) are new genes that enable an organism to adapt to its specific living environment. Our focus in this study is to compare ORFans between pathogens (P) and nonpathogens (NP) of the same genus. Using the pangenome idea, we have identified 130,169 ORFans in nine bacterial genera (505 genomes) and classified these ORFans into four groups: (i) SS-ORFans (P), which are only found in a single pathogenic genome; (ii) SS-ORFans (NP), which are only found in a single nonpathogenic genome; (iii) PS-ORFans (P), which are found in multiple pathogenic genomes; and (iv) NS-ORFans (NP), which are found in multiple nonpathogenic genomes. Within the same genus, pathogens do not always have more genes, more ORFans, or more pathogenicity-related genes (PRGs)-including prophages, pathogenicity islands (PAIs), virulence factors (VFs), and horizontal gene transfers (HGTs)-than nonpathogens. Interestingly, in pathogens of the nine genera, the percentages of PS-ORFans are consistently higher than those of SS-ORFans, which is not true in nonpathogens. Similarly, in pathogens of the nine genera, the percentages of PS-ORFans matching the four types of PRGs are also always higher than those of SS-ORFans, but this is not true in nonpathogens. All of these findings suggest the greater importance of PS-ORFans for bacterial pathogenicity. IMPORTANCE Recent pangenome analyses of numerous bacterial species have suggested that each genome of a single species may have a significant fraction of its gene content unique or shared by a very few genomes (i.e., ORFans). We selected nine bacterial genera, each containing at least five pathogenic and five nonpathogenic genomes, to compare their ORFans in relation to pathogenicity-related genes. Pathogens in these genera are known to cause a number of common and devastating human diseases such as pneumonia, diphtheria, melioidosis, and tuberculosis. Thus, they are worthy of in-depth systems microbiology investigations, including the comparative study of ORFans between pathogens and nonpathogens. We provide direct evidence to suggest that ORFans shared by more pathogens are more associated with pathogenicity-related genes and thus are more important targets for development of new diagnostic markers or therapeutic drugs for bacterial infectious diseases.
Keywords: ORFan; horizontal gene transfer; orphan gene; pathogenic island; pathogenicity; prophage; virulence factor.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
