epiCaPture: A Urine DNA Methylation Test for Early Detection of Aggressive Prostate Cancer
- PMID: 30801051
- PMCID: PMC6383793
- DOI: 10.1200/PO.18.00134
epiCaPture: A Urine DNA Methylation Test for Early Detection of Aggressive Prostate Cancer
Abstract
Purpose: Liquid biopsies that noninvasively detect molecular correlates of aggressive prostate cancer (PCa) could be used to triage patients, reducing the burdens of unnecessary invasive prostate biopsy and enabling early detection of high-risk disease. DNA hypermethylation is among the earliest and most frequent aberrations in PCa. We investigated the accuracy of a six-gene DNA methylation panel (Epigenetic Cancer of the Prostate Test in Urine [epiCaPture]) at detecting PCa, high-grade (Gleason score greater than or equal to 8) and high-risk (D'Amico and Cancer of the Prostate Risk Assessment] PCa from urine.
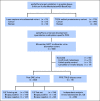
Patients and methods: Prognostic utility of epiCaPture genes was first validated in two independent prostate tissue cohorts. epiCaPture was assessed in a multicenter prospective study of 463 men undergoing prostate biopsy. epiCaPture was performed by quantitative methylation-specific polymerase chain reaction in DNA isolated from prebiopsy urine sediments and evaluated by receiver operating characteristic and decision curves (clinical benefit). The epiCaPture score was developed and validated on a two thirds training set to one third test set.
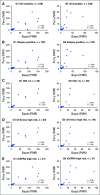
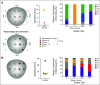
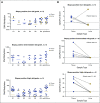
Results: Higher methylation of epiCaPture genes was significantly associated with increasing aggressiveness in PCa tissues. In urine, area under the receiver operating characteristic curve was 0.64, 0.86, and 0.83 for detecting PCa, high-grade PCa, and high-risk PCa, respectively. Decision curves revealed a net benefit across relevant threshold probabilities. Independent analysis of two epiCaPture genes in the same clinical cohort provided analytical validation. Parallel epiCaPture analysis in urine and matched biopsy cores showed added value of a liquid biopsy.
Conclusion: epiCaPture is a urine DNA methylation test for high-risk PCa. Its tumor specificity out-performs that of prostate-specific antigen (greater than 3 ng/mL). Used as an adjunct to prostate-specific antigen, epiCaPture could aid patient stratification to determine need for biopsy.
Conflict of interest statement
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Eve O’Reilly No relationship to disclose Alexandra V. Tuzova No relationship to disclose Anna L. Walsh No relationship to disclose Niamh M. Russell No relationship to disclose Odharnaith O’Brien No relationship to disclose Sarah Kelly No relationship to disclose Odharna Ni Dhomhnallain No relationship to disclose Liam DeBarra No relationship to disclose Connie M. Dale No relationship to disclose Rick Brugman No relationship to disclose Gavin Clarke No relationship to disclose Olivia Schmidt No relationship to disclose Shane O’Meachair No relationship to disclose Dattatraya Patil No relationship to disclose Kathryn L. Pellegrini No relationship to disclose Neil Fleshner Honoraria: Amgen, Janssen Oncology, Bayer, Sanofi, AbbVie, Ferring Pharmaceuticals, Astellas Medivation Consulting or Advisory Role: Hybridyne Health Research Funding: Ferring (Inst), Astellas Pharma (Inst), Janssen Oncology (Inst), Amgen (Inst), Nucleix (Inst), Progenix (Inst), Spectracure AB (Inst) Julia Garcia No relationship to disclose Fang Zhao No relationship to disclose Stephen Finn Honoraria: Roche Research Funding: Amgen (Inst) Travel, Accommodations, Expenses: Pfizer Robert Mills No relationship to disclose Marcelino Y. Hanna No relationship to disclose Rachel Hurst No relationship to disclose Elizabeth McEvoy No relationship to disclose William M. Gallagher Employment: OncoMark Leadership: OncoMark Stock and Other Ownership Interests: OncoMark Consulting or Advisory Role: Carrick Therapeutics Research Funding: Carrick Therapeutics Patents, Royalties, Other Intellectual Property: Two patents which have been licensed to OncoMark Travel, Accommodations, Expenses: OncoMark Rustom P. Manecksha Honoraria: Janssen, Boston Scientific Travel, Accommodations, Expenses: Ferring Pharmaceuticals, Astellas Pharma Colin S. Cooper Patents, Royalties, Other Intellectual Property: I have two patents for tests for aggressive prostate cancer filed in the past 2 years. These patents are not related to the current work. (Inst) Daniel S. Brewer Patents, Royalties, Other Intellectual Property: Patents held in drug discovery with Novartis (I), patents pending concerning cancer subtype detection and biomarkers with University of East Anglia Bharati Bapat No relationship to disclose Martin G. Sanda No relationship to disclose Jeremy Clark No relationship to disclose Antoinette S. Perry Patents, Royalties, Other Intellectual Property: University College Dublin holds a patent that relates to this work
Figures



References
-
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; http://globocan.iarc.fr
-
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. - PubMed
-
- Wilt TJ, Jones KM, Barry MJ, et al. Follow-up of prostatectomy versus observation for early prostate cancer. N Engl J Med. 2017;377:132–142. - PubMed
-
- Roobol MJ, Vedder MM, Nieboer D, et al. Comparison of two prostate cancer risk calculators that include the prostate health index. Eur Urol Focus. 2015;1:185–190. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

