Analytical Validation of a Single-nucleotide Polymorphism-based Donor-derived Cell-free DNA Assay for Detecting Rejection in Kidney Transplant Patients
- PMID: 30801536
- PMCID: PMC6867667
- DOI: 10.1097/TP.0000000000002665
Analytical Validation of a Single-nucleotide Polymorphism-based Donor-derived Cell-free DNA Assay for Detecting Rejection in Kidney Transplant Patients
Abstract
Background: Early detection of rejection in kidney transplant recipients holds the promise to improve clinical outcomes. Development and implementation of more accurate, noninvasive methods to detect allograft rejection remain an ongoing challenge. The limitations of existing allograft surveillance methods present an opportunity for donor-derived cell-free DNA (dd-cfDNA), which can accurately and rapidly differentiate patients with allograft rejection from patients with stable organ function.
Methods: This study evaluated the analytical performance of a massively multiplexed polymerase chain reaction assay that targets 13 962 single-nucleotide polymorphisms, characterized and validated using 66 unique samples with 1064 replicates, including cell line-derived reference samples, plasma-derived mixtures, and transplant patient samples. The dd-cfDNA fraction was quantified in both related and unrelated donor-recipient pairs.
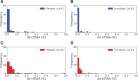
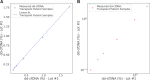
Results: The dd-cfDNA assay showed a limit of blank of 0.11%, a limit of detection and limit of quantitation of 0.15% for unrelated donors, and limit of blank of 0.23%, a limit of detection and limit of quantitation of 0.29% for related donors. All other metrics (linearity, accuracy, and precision) were observed to be equivalent between unrelated and related donors. The measurement precision of coefficient of variation was 1.8% (repeatability, 0.6% dd-cfDNA) and was <5% for all the different reproducibility measures.
Conclusions: This study validates the performance of a single-nucleotide polymorphism-based massively multiplexed polymerase chain reaction assay to detect the dd-cfDNA fraction with improved precision over currently available tests, regardless of donor-recipient relationships.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Comment in
-
Performance of Donor-derived Cell-free DNA Assays in Kidney Transplant Patients.Transplantation. 2020 May;104(5):e135. doi: 10.1097/TP.0000000000003084. Transplantation. 2020. PMID: 32044893 No abstract available.
-
The Author's Reply to "Performance of Donor-Derived Cell-Free DNA Assays in Kidney Transplant Patients".Transplantation. 2020 May;104(5):e136-e137. doi: 10.1097/TP.0000000000003085. Transplantation. 2020. PMID: 32317616 No abstract available.
References
-
- United States Renal Data System. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2018. 2018 USRDS annual data report: epidemiology of kidney disease in the United States. Available at https://www.usrds.org/2018/view/v2_06.aspx. Accessed December 12, 2018.
-
- Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn 201618890–902 - PubMed
-
- Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant 201111450–462 - PubMed
-
- Arnaud CH. Uncovering the hidden signs of organ transplant rejection. Chem Eng News. 2018;96:5.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

