A single test approach for accurate and sensitive detection and taxonomic characterization of Trypanosomes by comprehensive analysis of internal transcribed spacer 1 amplicons
- PMID: 30802245
- PMCID: PMC6414030
- DOI: 10.1371/journal.pntd.0006842
A single test approach for accurate and sensitive detection and taxonomic characterization of Trypanosomes by comprehensive analysis of internal transcribed spacer 1 amplicons
Abstract
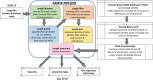
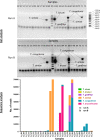
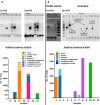
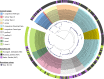
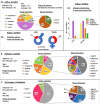
To improve our knowledge on the epidemiological status of African trypanosomiasis, better tools are required to monitor Trypanosome genotypes circulating in both mammalian hosts and tsetse fly vectors. This is important in determining the diversity of Trypanosomes and understanding how environmental factors and control efforts affect Trypanosome evolution. We present a single test approach for molecular detection of different Trypanosome species and subspecies using newly designed primers to amplify the Internal Transcribed Spacer 1 region of ribosomal RNA genes, coupled to Illumina sequencing of the amplicons. The protocol is based on Illumina's widely used 16s bacterial metagenomic analysis procedure that makes use of multiplex PCR and dual indexing. Results from analysis of wild tsetse flies collected from Zambia and Zimbabwe show that conventional methods for Trypanosome species detection based on band size comparisons on gels is not always able to accurately distinguish between T. vivax and T. godfreyi. Additionally, this approach shows increased sensitivity in the detection of Trypanosomes at species level with the exception of the Trypanozoon subgenus. We identified subspecies of T. congolense, T. simiae, T. vivax, and T. godfreyi without the need for additional tests. Results show T. congolense Kilifi subspecies is more closely related to T. simiae than to other T. congolense subspecies. This agrees with previous studies using satellite DNA and 18s RNA analysis. While current classification does not list any subspecies for T. godfreyi, we observed two distinct clusters for these species. Interestingly, sequences matching T. congolense Tsavo (now classified as T. simiae Tsavo) clusters distinctly from other T. simiae Tsavo sequences suggesting the Nannomonas group is more divergent than currently thought thus the need for better classification criteria. This method presents a simple but comprehensive way of identification of Trypanosome species and subspecies-specific using one PCR assay for molecular epidemiology of trypanosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Molecular identification of different trypanosome species and subspecies in tsetse flies of northern Nigeria.Parasit Vectors. 2016 May 23;9(1):301. doi: 10.1186/s13071-016-1585-3. Parasit Vectors. 2016. PMID: 27216812 Free PMC article.
-
Evaluation of ITS1 rDNA primers for the detection and identification of African trypanosomes in mammalian hosts and tsetse flies.Acta Trop. 2024 Oct;258:107331. doi: 10.1016/j.actatropica.2024.107331. Epub 2024 Jul 24. Acta Trop. 2024. PMID: 39059714
-
The use of molecular technology to investigate trypanosome infections in tsetse flies at Liwonde Wild Life Reserve.Malawi Med J. 2019 Dec;31(4):233-237. doi: 10.4314/mmj.v31i4.3. Malawi Med J. 2019. PMID: 32133052 Free PMC article.
-
Species-specific probes for the identification of the African tsetse-transmitted trypanosomes.Parasitology. 2009 Oct;136(12):1501-7. doi: 10.1017/S0031182009006179. Epub 2009 Jun 2. Parasitology. 2009. PMID: 19490726 Review.
-
Landmarks in the evolution of technologies for identifying trypanosomes in tsetse flies.Trends Parasitol. 2010 Aug;26(8):388-94. doi: 10.1016/j.pt.2010.04.011. Epub 2010 Jun 9. Trends Parasitol. 2010. PMID: 20542733 Review.
Cited by
-
Comprehensive analysis of oral administration of Vitamin E on the early stage of Trypanosoma brucei brucei infection.J Parasit Dis. 2021 Jun;45(2):512-523. doi: 10.1007/s12639-020-01322-5. Epub 2021 Jan 3. J Parasit Dis. 2021. PMID: 34295050 Free PMC article.
-
Molecular and genetic diversity in isolates of Trypanosoma evansi from naturally infected horse and dogs by using RoTat 1.2 VSG gene in Madhya Pradesh, India.Mol Biol Rep. 2023 Sep;50(9):7347-7356. doi: 10.1007/s11033-023-08651-7. Epub 2023 Jul 13. Mol Biol Rep. 2023. PMID: 37439897
-
Bovine Piroplasma Populations in the Philippines Characterized Using Targeted Amplicon Deep Sequencing.Microorganisms. 2023 Oct 18;11(10):2584. doi: 10.3390/microorganisms11102584. Microorganisms. 2023. PMID: 37894242 Free PMC article.
-
Molecular identification of new Trypanosoma evansi type non-A/B isolates from buffaloes and cattle in Indonesia.Rev Bras Parasitol Vet. 2024 Jun 28;33(2):e001324. doi: 10.1590/S1984-29612024033. eCollection 2024. Rev Bras Parasitol Vet. 2024. PMID: 38958293 Free PMC article.
-
Diversity of trypanosomes in wildlife of the Kafue ecosystem, Zambia.Int J Parasitol Parasites Wildl. 2020 Apr 23;12:34-41. doi: 10.1016/j.ijppaw.2020.04.005. eCollection 2020 Aug. Int J Parasitol Parasites Wildl. 2020. PMID: 32420023 Free PMC article.
References
-
- WHO. Investing to overcome the global impact of neglected tropical diseases: third WHO report on neglected diseases 2015. Invest to overcome Glob impact neglected Trop Dis third WHO Rep neglected Dis. 2015; 191. ISBN 978 92 4 156486 1
-
- Médecins SF. SLEEPING SICKNESS or Human African Trypanosomiasis Fact sheet [Internet]. 2004 [cited 27 Feb 2018]. Available: http://www.accessmed-msf.org/fileadmin/user_upload/diseases/other_diseas...
-
- Levine ND. The Trypanosomes of Mammals. A Zoological Monograph. Cecil A. Hoare. Blackwell, Oxford, England, 1972 (U.S. distributor, Davis, Philadelphia). xviii, 750 pp. + plates. Science (80-). 1973;179: 60. Available: http://science.sciencemag.org/content/179/4068/60.abstract
-
- World Health Organization. Control and surveillance of human African trypanosomiasis. World Health Organ Tech Rep Ser. 2013; 1–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

