PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery
- PMID: 30802273
- PMCID: PMC6405170
- DOI: 10.1371/journal.ppat.1007590
PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery
Abstract
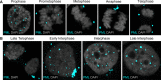
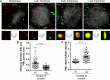
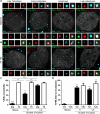
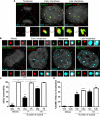
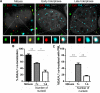
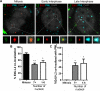
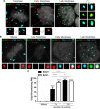
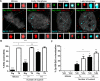
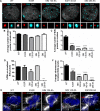
Subnuclear promyelocytic leukemia (PML) nuclear bodies (NBs) are targeted by many DNA viruses after nuclear delivery. PML protein is essential for formation of PML NBs. Sp100 and Small Ubiquitin-Like Modifier (SUMO) are also permanently residing within PML NBs. Often, large DNA viruses disassemble and reorganize PML NBs to counteract their intrinsic antiviral activity and support establishment of infection. However, human papillomavirus (HPV) requires PML protein to retain incoming viral DNA in the nucleus for subsequent efficient transcription. In contrast, Sp100 was identified as a restriction factor for HPV. These findings suggested that PML NBs are important regulators of early stages of the HPV life cycle. Nuclear delivery of incoming HPV DNA requires mitosis. Viral particles are retained within membrane-bound transport vesicles throughout mitosis. The viral genome is released from transport vesicles by an unknown mechanism several hours after nuclear envelope reformation. The minor capsid protein L2 mediates intracellular transport by becoming transmembranous in the endocytic compartment. Herein, we tested our hypothesis that PML protein is recruited to incoming viral genome prior to egress from transport vesicles. High-resolution microscopy revealed that PML protein, SUMO-1, and Sp100 are recruited to incoming viral genomes, rather than viral genomes being targeted to preformed PML NBs. Differential immunofluorescent staining suggested that PML protein and SUMO-1 associated with transport vesicles containing viral particles prior to egress, implying that recruitment is likely mediated by L2 protein. In contrast, Sp100 recruitment to HPV-harboring PML NBs occurred after release of viral genomes from transport vesicles. The delayed recruitment of Sp100 is specific for HPV-associated PML NBs. These data suggest that the virus continuously resides within a protective environment until the transport vesicle breaks down in late G1 phase and imply that HPV might modulate PML NB assembly to achieve establishment of infection and the shift to viral maintenance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95(9):2748–52. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

