lncINS-IGF2 Promotes Cell Proliferation and Migration by Promoting G1/S Transition in Lung Cancer
- PMID: 30803359
- PMCID: PMC6374000
- DOI: 10.1177/1533033818823029
lncINS-IGF2 Promotes Cell Proliferation and Migration by Promoting G1/S Transition in Lung Cancer
Abstract
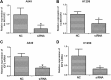
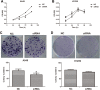
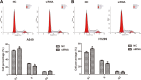
Long noncoding RNAs are capable of regulating gene expression at multiple levels. These RNA molecules are also involved in a variety of physiological and pathological processes. Emerging data demonstrate that a series of differentially expressed long noncoding RNAs are implicated in tumorigenesis. In the present study, we used microarray analysis to identify long noncoding RNAs that are dysregulated in non-small-cell lung cancer when compared to normal lung tissues. Accordingly, we performed quantitative real-time polymerase chain reaction to analyze the levels of long noncoding RNA and the cis target gene. We further found the oncogene property of long noncoding RNA that long noncoding RNA downexpression inhibits non-small-cell lung cancer cells proliferation and migration based on 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide and colony formation assays and wound healing as well as transwell assays. The influence of long noncoding RNA on cell cycle of non-small-cell lung cancer cells is also analyzed by flow cytometry. Among the dysregulated long noncoding RNAs, we identified INS-IGF2 readthrough, transcript variant 1, noncoding RNA (NR_003512.3) is upregulated in non-small-cell lung cancer tissues, the cis gene of which is insulin-like growth factor 2 gene hinted by bioinformatics analysis. We also observed that downregulation of INS-IGF2 readthrough, transcript variant 1, noncoding RNA reduces insulin-like growth factor 2 messenger RNA expression. Furthermore, INS-IGF2 readthrough, transcript variant 1, noncoding RNA downregulation suppresses non-small-cell lung cancer cell proliferation and migration. This downregulation results in a concomitant inhibition of the G1/S transition in non-small-cell lung cancer cells. Our findings suggest that INS-IGF2 readthrough, transcript variant 1, noncoding RNA may be an oncogene involved in the development of lung cancer. Therefore, we speculate that INS-IGF2 readthrough, transcript variant 1, noncoding RNA represents a potential therapeutic target for lung cancer.
Keywords: lncINS-IGF2; lncRNA; lung cancer; migration; proliferation.
Conflict of interest statement
Figures
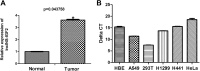
Similar articles
-
Long non-coding RNA FENDRR inhibits NSCLC cell growth and aggressiveness by sponging miR-761.Eur Rev Med Pharmacol Sci. 2018 Dec;22(23):8324-8332. doi: 10.26355/eurrev_201812_16530. Eur Rev Med Pharmacol Sci. 2018. PMID: 30556873
-
Long noncoding RNA HEIH promotes the proliferation and metastasis of non-small cell lung cancer.J Cell Biochem. 2019 Mar;120(3):3529-3538. doi: 10.1002/jcb.27629. Epub 2018 Sep 19. J Cell Biochem. 2019. PMID: 30230600
-
LncRNA SNHG1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the miR-361-3p/FRAT1 axis.Thorac Cancer. 2020 Feb;11(2):295-304. doi: 10.1111/1759-7714.13256. Epub 2019 Dec 2. Thorac Cancer. 2020. PMID: 31788970 Free PMC article.
-
Emerging roles of long noncoding RNA H19 in human lung cancer.Cell Biochem Funct. 2024 Jun;42(4):e4072. doi: 10.1002/cbf.4072. Cell Biochem Funct. 2024. PMID: 39031589 Review.
-
[Long non-coding RNAs, new therapeutic targets for the treatment of non-small cell lung cancers?].Rev Mal Respir. 2025 Mar;42(3):143-147. doi: 10.1016/j.rmr.2025.02.002. Epub 2025 Mar 10. Rev Mal Respir. 2025. PMID: 40069040 Review. French.
Cited by
-
Comprehensive analysis of competing endogenous RNA network in Wilms tumor based on the TARGET database.Transl Androl Urol. 2020 Apr;9(2):473-484. doi: 10.21037/tau.2020.01.34. Transl Androl Urol. 2020. PMID: 32420153 Free PMC article.
-
The metabolic effects of resumption of a high fat diet after weight loss are sex dependent in mice.Sci Rep. 2023 Aug 14;13(1):13227. doi: 10.1038/s41598-023-40514-w. Sci Rep. 2023. PMID: 37580448 Free PMC article.
-
Altered methylation of imprinted genes in neuroblastoma: implications for prognostic refinement.J Transl Med. 2024 Aug 31;22(1):808. doi: 10.1186/s12967-024-05634-5. J Transl Med. 2024. PMID: 39217334 Free PMC article.
-
Phaeochromocytomas overexpress insulin transcript and produce insulin.Endocr Connect. 2021 Jul 26;10(8):815-824. doi: 10.1530/EC-21-0269. Endocr Connect. 2021. PMID: 34170845 Free PMC article.
-
Fusion Transcripts of Adjacent Genes: New Insights into the World of Human Complex Transcripts in Cancer.Int J Mol Sci. 2019 Oct 23;20(21):5252. doi: 10.3390/ijms20215252. Int J Mol Sci. 2019. PMID: 31652751 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2014;64(1):9–29. - PubMed
-
- Seve P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. - PubMed
-
- Raungrut P, Wongkotsila A, Lirdprapamongkol K, et al. Prognostic significance of 14-3-3gamma overexpression in advanced non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15(8):3513–3518. - PubMed
-
- Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. J Genet. 2015;94(4):771–784. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

