Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development
- PMID: 30803449
- PMCID: PMC6388476
- DOI: 10.1186/s13073-019-0621-2
Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development
Abstract
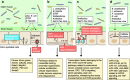
In recent years, the number of studies investigating the impact of the gut microbiome in colorectal cancer (CRC) has risen sharply. As a result, we now know that various microbes (and microbial communities) are found more frequently in the stool and mucosa of individuals with CRC than healthy controls, including in the primary tumors themselves, and even in distant metastases. We also know that these microbes induce tumors in various mouse models, but we know little about how they impact colon epithelial cells (CECs) directly, or about how these interactions might lead to modifications at the genetic and epigenetic levels that trigger and propagate tumor growth. Rates of CRC are increasing in younger individuals, and CRC remains the second most frequent cause of cancer-related deaths globally. Hence, a more in-depth understanding of the role that gut microbes play in CRC is needed. Here, we review recent advances in understanding the impact of gut microbes on the genome and epigenome of CECs, as it relates to CRC. Overall, numerous studies in the past few years have definitively shown that gut microbes exert distinct impacts on DNA damage, DNA methylation, chromatin structure and non-coding RNA expression in CECs. Some of the genes and pathways that are altered by gut microbes relate to CRC development, particularly those involved in cell proliferation and WNT signaling. We need to implement more standardized analysis strategies, collate data from multiple studies, and utilize CRC mouse models to better assess these effects, understand their functional relevance, and leverage this information to improve patient care.
Conflict of interest statement
Competing interests
CLS is supported, in part, by a grant from Bristol-Myers Squibb; JA declares that he has no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

