Cytoplasmic control of Rab family small GTPases through BAG6
- PMID: 30804014
- PMCID: PMC6446207
- DOI: 10.15252/embr.201846794
Cytoplasmic control of Rab family small GTPases through BAG6
Abstract
Rab family small GTPases are master regulators of distinct steps of intracellular vesicle trafficking in eukaryotic cells. GDP-bound cytoplasmic forms of Rab proteins are prone to aggregation due to the exposure of hydrophobic groups but the machinery that determines the fate of Rab species in the cytosol has not been elucidated in detail. In this study, we find that BAG6 (BAT3/Scythe) predominantly recognizes a cryptic portion of GDP-associated Rab8a, while its major GTP-bound active form is not recognized. The hydrophobic residues of the Switch I region of Rab8a are essential for its interaction with BAG6 and the degradation of GDP-Rab8a via the ubiquitin-proteasome system. BAG6 prevents the excess accumulation of inactive Rab8a, whose accumulation impairs intracellular membrane trafficking. BAG6 binds not only Rab8a but also a functionally distinct set of Rab family proteins, and is also required for the correct distribution of Golgi and endosomal markers. From these observations, we suggest that Rab proteins represent a novel set of substrates for BAG6, and the BAG6-mediated pathway is associated with the regulation of membrane vesicle trafficking events in mammalian cells.
Keywords: BAG6; Rab GTP‐binding proteins; Rab8a; membrane trafficking; ubiquitin‐proteasome system.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
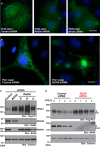
- A
At 72 h after transfection with siRNA duplexes for BAG6 or control siRNA (10 nM each), the intracellular localization of TfnR in HeLa cells was examined (shown as green). Nuclear DNA was stained with Hoechst 33342 (shown as blue). Control knockdown (left panel), Rab8a knockdown (center panel), and BAG6 knockdown (right panel). Efficacy of endogenous BAG6 knockdown in HeLa cells was verified by Western blot experiments (see Fig EV1D). Scale bar: 10 μm.
- B
Intracellular localization of Ptc1 (green) in HeLa cells. Nuclei were stained with Hoechst 33342 (shown as blue). See also Fig EV1A and B. Scale bar: 10 μm.
- C, D
Knockdown of Rab8a (with Rab8a siRNA#1, #2, and #3) or BAG6 (with BAG6 siRNA#1) stimulated the accumulation and stabilization of Ptc1 protein in HEK293 cells. See also Appendix Fig S1B.

The abnormal Ptc1 signal observed in BAG6‐suppressed HeLa cells at the perinuclear region was not derived from protein aggregates. The Ptc1 immunosignal in BAG6‐suppressed cells (shown as magenta with an arrowhead) was negative for FK2 polyubiquitin staining (shown as green), a marker for cytoplasmic protein aggregates. See also Appendix Fig S1A. Nuclei were stained by Hoechst 33342 (shown as blue). Scale bar: 10 μm.
Immunostaining of Ptc1 (green) in HeLa cells that were treated with or without Rab8a siRNA. See also Appendix Fig S1D. Scale bar: 10 μm.
Immunostaining of the ER luminal marker protein calnexin (green) in HeLa cells that were treated with or without BAG6 siRNA. Scale bar: 5 μm.
Cell lysates were subjected to Western blot analysis with an anti‐BAG6 antibody to verify the knockdown efficacy of BAG6 siRNA#1, #2, and #3 in HeLa cells. As a negative control, BAG6 siRNA#1scr was used. Actin was used as a loading control.

Protein interaction network suggested by public databases.
BAG6 protein co‐precipitated Rab8a, while neither Rab7 nor Rab11a was co‐precipitated with BAG6. Flag‐tagged Rab8a, Rab7, Rab11a, and luciferase‐CL1 (Lc‐CL1; a positive control) were expressed in HeLa cells and the cells were treated with 10 μM MG‐132 for 4 h. Flag‐immunoprecipitates were blotted with anti‐BAG6 and anti‐Flag antibodies, respectively. Note that all cells used were treated with 10 μM MG‐132 for 4 h.
S‐tagged BAG6 pull‐down efficiently co‐precipitated Rab8a (T22N), a GDP‐bound mutant, while BAG6 scarcely co‐precipitated Rab8a (Q67L), a constitutively active mutant. Co‐precipitation of Rab8a WT with BAG6 was used as a standard. MG‐132 (10 μM) was included in the cell culture for 4 h, as indicated. Note that the T22N mutant protein was expressed at lower levels than either WT or Q67L, and that this was partly due to increased degradation, as will be shown later. S‐BAG6 stands for N‐terminally S‐tagged BAG6 protein (see the Materials and Methods).
Anti‐Flag signals in (C) were quantified, and relative signal intensities are presented. The value of the WT Rab8a signal with MG‐132 was defined as 1.0. Note that all signal intensities of Flag‐tag were normalized by that of the Rab8a input signals. The graph represents the mean ± standard deviation (SD) calculated from three independent biological replicates. An asterisk indicates P < 0.05 (Student's t‐test).
A series of Flag‐Rab8a mutants were immunoprecipitated and quantified the amount of endogenous BAG6 that were co‐precipitated with Flag‐Rab8a. Note that all cells used were treated with 10 μM MG‐132 for 4 h.
Deficiency of Rabin8, a GEF for Rab8a, enhanced the physical interaction between BAG6 and WT Rab8a proteins. Flag‐tagged WT Rab8a was expressed in Rabin8 siRNA‐treated cells, and Flag‐immunoprecipitates were probed with an anti‐BAG6 antibody. Note that all cells used were treated with 10 μM MG‐132 for 4 h.

- A
Endogenous Rab8a protein was co‐precipitated with S‐tagged BAG6 N465 from HeLa cell lysate in the presence of MG‐132.
- B
Co‐immunoprecipitation of BAG6 with Rab8a was stimulated by the addition of a proteasome inhibitor. HeLa cells expressing Flag‐tagged Rab8a (T22N) protein were treated with (+) or without (−) 10 μM MG‐132. At 4 h after MG‐132 treatment, the cells were lysed and Flag‐precipitates were probed with an anti‐BAG6 antibody.
- C
Rab8a C204S mutant protein localized exclusively in the soluble cytoplasmic fraction. Whole cell extracts of HeLa cells expressing Flag‐tagged Rab8a proteins (WT or C204S mutant) were fractionated into the cytosolic and membrane fractions. Tubulin was used as a cytoplasmic marker.
- D, E
T22N mutant form of full‐length Rab8a protein was stabilized by MG‐132, while leupeptin did not affect its stability. At 24 h after Rab8a transfection, the cells were cultured with 10 μM MG‐132, 10 μM leupeptin, or an equivalent amount of DMSO (as a negative control), and then chased with 20 μg/ml CHX and harvested at the indicated times after CHX addition.
- F, G
Anti‐Flag immunosignals of Rab8a WT, T22N, Q67L, and T22N‐3IS proteins in Figs 3E and 4D were quantified. The data represent the mean ± SD calculated from three independent biological replicates (n = 3). *P < 0.05 compared with control siRNA. N.S. indicates not significant (Student's t‐test).

- A
Schematic representation of Rab8a protein. To prevent C‐terminal geranylgeranylation, the Cys204 residue was substituted with a serine residue. Alternatively, two truncated proteins (N100 and ∆N100 fragments of Rab8a) were prepared for this experiment.
- B
C‐terminal geranylgeranylation of Rab8a was dispensable for BAG6 recognition.
- C
The N‐terminal GTPase domain of Rab8a was essential for BAG6 recognition. A series of Flag‐tagged truncated fragments of WT Rab8a were expressed in HeLa cells with S‐tagged BAG6 and treated with (+) or without (−) protease inhibitors for 4 h. Flag‐Rab8a substrates were immunoprecipitated and probed with an anti‐BAG6 antibody.
- D
Hydrophobicity recognition domain of BAG6 (N465) was critical for Rab8a recognition. A schematic representation of the BAG6‐truncated proteins used in this experiment is shown in the upper panel. Numbers denote the corresponding amino acids of mammalian BAG6. Positions of UBL, BUILD, and DUF3538 domains, which are all linked to hydrophobicity recognition by BAG6 37, are indicated. WT, wild‐type; ΔN465, N‐terminal 465 residues‐deleted mutant; and Ν465, C‐terminal 689 residues‐deleted mutant.
- E, F
CHX chase experiments show that GDP‐bound Rab8a (T22N) was a highly labile protein, while the GTP‐bound active mutant (Q67L) was a stable protein (E). Instability of the T22N mutant was not perturbed by the C204S mutation (F). Actin was used as a loading control.
- G
N‐terminal 100 residue fragment of the Rab8a GTPase domain was sensitive to the proteasome inhibitor, while the ∆N100 fragment was not.
- H
Rab8a (T22N) was polyubiquitinated in the presence of the proteasome inhibitor. Asterisks indicate non‐specific bands.

Schematic representation of the two Switch regions (I and II) within the Rab8a GTPase domain. Numbers denote the corresponding amino acids of human Rab8a (upper panel). Amino acid sequence alignments of the Switch I region of Rab family proteins (lower panel). Three conserved hydrophobic residues (Ile38, Ile41, and Ile43) in this region are indicated in red, while the other hydrophobic residues are indicated in orange. The three hydrophobic residues were substituted with serine and this construct was designated as the T22N‐3IS mutant. Note that Ile38 and Ile43 of Rab8a are not conserved in Rab7.
Kyte‐Doolittle hydrophobicity plots of the complete amino acid sequence of human WT Rab8a and T22N‐3IS mutant. The hydrophobicity peak within WT Switch I was abolished in the 3IS mutant (indicated within the red box). The numbers on the horizontal axis denote the corresponding amino acid positions in these proteins.
Substitution of the hydrophobic residues of Switch I with hydrophilic residues (T22N 3IS) abolished its binding to BAG6 protein.
Rab8a (T22N‐3IS) mutant protein was highly stable, while Rab8a (T22N) protein was quite unstable in HeLa cells. Actin was used as a loading control.
Rab8a (T22N‐3IS) mutant was not subject to polyubiquitin co‐precipitation, while Rab8a (T22N) protein was.

Schematic representation of the Switch I region of the Rab8a‐Rab7a chimeric protein. The amino acid residues 33–45 (boxed region) of full‐length Rab8a were substituted with those of Rab7a and this mutant protein was designated as the “8a‐7a chimera”. The numbers denote the corresponding amino acids of human Rab8a and Rab7a.
The 8a‐7a chimera protein with the T22N mutation showed greatly reduced affinity with BAG6 compared with the case in Rab8a (T22N).

Rab8a (T22N) protein accumulated in BAG6‐knockdown cells. HeLa cells were transfected with siRNA duplexes for BAG6 or control siRNA. At 48 h after siRNA transfection, Flag‐tagged‐Rab8a (T22N) was expressed in the cells. At 24 h after Rab8a (T22N) transfection, the cells were chased with 50 μg/ml CHX and harvested at the indicated time after CHX addition. Actin was used as a loading control.
Anti‐Flag blot signals in the control or BAG6 siRNA‐treated cells were quantified, and relative signal intensities after CHX addition were calculated. The value of the Flag‐signal at 0 h was defined as 1.0. Note that all signal intensities of the Flag‐tag were normalized by that of actin, a loading control, in each sample. The graph represents the mean ± SE calculated from six independent biological replicates. These data were analyzed by Welch's t‐test.
Polyubiquitin modification of Rab8a was abolished in BAG6‐knockdown cells. Flag‐Rab8a (T22N) immunoprecipitates were blotted with an anti‐polyubiquitin antibody (FK2, left panel). As a negative control, siRNA#1scr was used. Anti‐polyubiquitin signals co‐precipitated with Rab8a (T22N) (a representative example is shown in the left panel) were quantified (right panel). Note that the intensities of the co‐precipitated polyubiquitin signal were normalized both by the input ubiquitin‐signal and bait Flag‐signal. The graph represents the mean ± SD calculated from three independent biological replicates. An asterisk indicates P < 0.05 (Student's t‐test).
Defective distribution of the endosomal protein TfnR in HeLa cells with the excess accumulation of the inactive form of Rab8a. Right panel shows a merged image of TfnR staining (shown as green), Rab8a (T22N) staining (magenta), and Hoechst 33342 nuclear staining (blue). Scale bar: 10 μm.
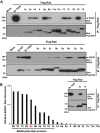
A series of major Rab family proteins were immunoprecipitated from HeLa cell extracts and then probed with an anti‐BAG6 antibody. Co‐precipitation of Rab8a (T22N) with BAG6 was used as a positive control for this experiment.
Quantification of BAG6 co‐precipitation with Rab family proteins. The graph shows the quantities of BAG6 protein in the respective anti‐Flag precipitates that were normalized by the amount of Flag‐Rab bait proteins. The value of BAG6 co‐precipitation with Rab8a (T22N) was defined as 1.0. The quantities of the respective immunosignals were also normalized to the actin signal of each sample and represent the median calculated from at least three independent biological replicates. The number of independent biological replicates was as follows: Rab1a, n = 4; Rab1b, n = 5; Rab2b, n = 3; Rab3a, n = 4; Rab4, n = 4; Rab5a, n = 3; Rab5b, n = 4; Rab6a, n = 5; Rab6b, n = 4; Rab7a, n = 3; Rab8a, n = 14; Rab9b, n = 4; Rab10, n = 6; Rab11a, n = 3; Rab11b, n = 3; Rab12, n = 2; Rab13, n = 6; Rab14, n = 3; Rab15, n = 5; Rab35, n = 3.

- A, B
BAG6 knockdown induced the abnormal distribution of Golgi apparatus markers. Representative images of the trans‐Golgi membrane protein Stx6 (green) in BAG6‐suppressed CHO cells with a Chinese hamster‐specific siRNA (cBAG6 siRNA#2). Scale bar: 10 μm (A). Images of the cis‐Golgi membrane protein GS28 and the cis‐Golgi matrix protein GM130 in BAG6‐suppressed CHO cells with another Chinese hamster‐specific siRNA (cBAG6 siRNA#5). GS28 (green) and GM130 (red) are indicated by arrowheads. Scale bar: 10 μm. (B). Fluorescent signals were detected using a laser scanning confocal microscopy system. Nuclei were stained by Hoechst (blue).
- C
Glycosylation of the IL‐2Rα transmembrane protein was not reduced by BAG6 knockdown. Flag‐tagged WT IL‐2Rα protein was expressed in HeLa cells with (+) or without (−) BAG6 siRNA, and was immunoprecipitated with an anti‐Flag antibody. The precipitates were incubated with (+) or without (−) 10 unit of the deglycosylation enzyme PNGase F and subjected to Western blot analysis with an anti‐Flag antibody. Low‐mobility (indicated as glycosylated) and high‐mobility (indicated as non‐glycosylated) signals of WT IL‐2Rα are indicated.
- D–F
Defects in the distribution of cell surface glycoproteins in BAG6‐suppressed cells. Representative image from a cell surface glycoprotein quantification assay with Alexa Fluor™ 488‐conjugated Lectin GS‐II as a probe (D). The graph quantitatively displays the number of fluorescence counts per cell as the mean ± SD calculated from 10 independent biological replicates (E). Lectin GS‐II‐derived cell surface signals were counted using ImageJ software. SRP54 knockdown was used as a positive control for this experiment (see also Appendix Fig S6). *P < 0.05 compared with control knockdown (Student's t‐test). Cell surface fluorescent signals were detected by confocal microscopy without plasma membrane permeabilization. BAG6 siRNA down‐regulated the cell surface expression of glycoproteins (F).

Verification of the depletion efficacy and specificity of two independent double‐stranded RNAs (cBAG6 siRNA#2 and #5) in rodent CHO cells. Note that the target sequences of these rodent cBAG6 siRNAs are completely different to those of the human siRNAs used in Fig 1 (see Materials and Methods). Two scrambled sequences for cBAG6 siRNA#2 (designated #2scr‐1 and #2scr‐2, respectively), as well as MISSION siRNA Universal Negative Control 1 (indicated by a black dot), were used as negative controls. Efficacy of Rab8a knockdown in CHO cells was also verified by a blot with endogenous proteins.
Comparison of the defects observed in the distribution of Stx6 (green) with siRNAs (cBAG6 siRNA#5 and Rab8a siRNA#1, respectively) in CHO cells. Note that both cBAG6 siRNA#5 and #2 are Chinese hamster‐specific, while the target sequence of Rab8a siRNA#1 is identical between humans and hamsters. An asterisk indicates a non‐specific signal. Scale bar: 10 μm.
Comparison of the defects observed in the distribution of the ER‐Golgi SNARE protein GS28 (green) and the cis‐Golgi marker GM130 (red) with siRNAs (cBAG6 siRNA#2 or RAB8a siRNA#1 and their combination) in CHO cells. GS28 and GM130 signals were dispersed throughout the perinuclear region of the cytoplasm in Rab8a knockdown cells, a similar phenotype to that observed in BAG6‐depleted cells. Scale bar: 10 μm.

Similar articles
-
BAG6 contributes to glucose uptake by supporting the cell surface translocation of the glucose transporter GLUT4.Biol Open. 2020 Jan 24;9(1):bio047324. doi: 10.1242/bio.047324. Biol Open. 2020. PMID: 31911483 Free PMC article.
-
A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation.J Biol Chem. 2013 Jun 21;288(25):18068-76. doi: 10.1074/jbc.M112.449199. Epub 2013 May 12. J Biol Chem. 2013. PMID: 23665563 Free PMC article.
-
Rab41 is a novel regulator of Golgi apparatus organization that is needed for ER-to-Golgi trafficking and cell growth.PLoS One. 2013 Aug 6;8(8):e71886. doi: 10.1371/journal.pone.0071886. Print 2013. PLoS One. 2013. PMID: 23936529 Free PMC article.
-
BAG6/BAT3: emerging roles in quality control for nascent polypeptides.J Biochem. 2013 Feb;153(2):147-60. doi: 10.1093/jb/mvs149. Epub 2012 Dec 28. J Biochem. 2013. PMID: 23275523 Review.
-
Multiple Roles of Rab GTPases at the Golgi.Results Probl Cell Differ. 2019;67:95-123. doi: 10.1007/978-3-030-23173-6_6. Results Probl Cell Differ. 2019. PMID: 31435794 Review.
Cited by
-
GDP-bound Rab37 modulates M2-like tumor-associated macrophage polarization by attenuating STAT1 translocation to downregulate the type I IFN pathway.Br J Cancer. 2025 Apr;132(7):622-634. doi: 10.1038/s41416-025-02955-0. Epub 2025 Feb 21. Br J Cancer. 2025. PMID: 39984679
-
BAG6 contributes to glucose uptake by supporting the cell surface translocation of the glucose transporter GLUT4.Biol Open. 2020 Jan 24;9(1):bio047324. doi: 10.1242/bio.047324. Biol Open. 2020. PMID: 31911483 Free PMC article.
-
Protein quality control machinery supports primary ciliogenesis by eliminating GDP-bound Rab8-family GTPases.iScience. 2023 Apr 10;26(5):106652. doi: 10.1016/j.isci.2023.106652. eCollection 2023 May 19. iScience. 2023. PMID: 37182096 Free PMC article.
-
GDP/GTP exchange factor MADD drives activation and recruitment of secretory Rab GTPases to Weibel-Palade bodies.Blood Adv. 2021 Dec 14;5(23):5116-5127. doi: 10.1182/bloodadvances.2021004827. Blood Adv. 2021. PMID: 34551092 Free PMC article.
-
Rab family of small GTPases: an updated view on their regulation and functions.FEBS J. 2021 Jan;288(1):36-55. doi: 10.1111/febs.15453. Epub 2020 Jul 1. FEBS J. 2021. PMID: 32542850 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

