α-Difluoromethylornithine reduces gastric carcinogenesis by causing mutations in Helicobacter pylori cagY
- PMID: 30804204
- PMCID: PMC6421409
- DOI: 10.1073/pnas.1814497116
α-Difluoromethylornithine reduces gastric carcinogenesis by causing mutations in Helicobacter pylori cagY
Abstract
Infection by Helicobacter pylori is the primary cause of gastric adenocarcinoma. The most potent H. pylori virulence factor is cytotoxin-associated gene A (CagA), which is translocated by a type 4 secretion system (T4SS) into gastric epithelial cells and activates oncogenic signaling pathways. The gene cagY encodes for a key component of the T4SS and can undergo gene rearrangements. We have shown that the cancer chemopreventive agent α-difluoromethylornithine (DFMO), known to inhibit the enzyme ornithine decarboxylase, reduces H. pylori-mediated gastric cancer incidence in Mongolian gerbils. In the present study, we questioned whether DFMO might directly affect H. pylori pathogenicity. We show that H. pylori output strains isolated from gerbils treated with DFMO exhibit reduced ability to translocate CagA in gastric epithelial cells. Further, we frequently detected genomic modifications in the middle repeat region of the cagY gene of output strains from DFMO-treated animals, which were associated with alterations in the CagY protein. Gerbils did not develop carcinoma when infected with a DFMO output strain containing rearranged cagY or the parental strain in which the wild-type cagY was replaced by cagY with DFMO-induced rearrangements. Lastly, we demonstrate that in vitro treatment of H. pylori by DFMO induces oxidative DNA damage, expression of the DNA repair enzyme MutS2, and mutations in cagY, demonstrating that DFMO directly affects genomic stability. Deletion of mutS2 abrogated the ability of DFMO to induce cagY rearrangements directly. In conclusion, DFMO-induced oxidative stress in H. pylori leads to genomic alterations and attenuates virulence.
Keywords: Helicobacter pylori; chemoprevention; difluoromethylornithine; gastric cancer; polyamines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


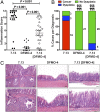
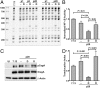
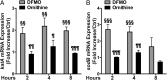

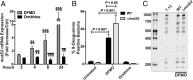
Similar articles
-
Helicobacter pylori CagA and Cag type IV secretion system activity have key roles in triggering gastric transcriptional and proteomic alterations.Infect Immun. 2025 Apr 8;93(4):e0059524. doi: 10.1128/iai.00595-24. Epub 2025 Mar 6. Infect Immun. 2025. PMID: 40047510 Free PMC article.
-
Genetic Manipulation of Helicobacter pylori Virulence Function by Host Carcinogenic Phenotypes.Cancer Res. 2017 May 1;77(9):2401-2412. doi: 10.1158/0008-5472.CAN-16-2922. Epub 2017 Feb 16. Cancer Res. 2017. PMID: 28209611 Free PMC article.
-
Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils.Gastroenterology. 2005 May;128(5):1229-42. doi: 10.1053/j.gastro.2005.02.064. Gastroenterology. 2005. PMID: 15887107
-
Clinical relevance of Helicobacter pylori vacA and cagA genotypes in gastric carcinoma.Best Pract Res Clin Gastroenterol. 2014 Dec;28(6):1003-15. doi: 10.1016/j.bpg.2014.09.004. Epub 2014 Oct 2. Best Pract Res Clin Gastroenterol. 2014. PMID: 25439067 Review.
-
Host genetic factors respond to pathogenic step-specific virulence factors of Helicobacter pylori in gastric carcinogenesis.Mutat Res Rev Mutat Res. 2014 Jan-Mar;759:14-26. doi: 10.1016/j.mrrev.2013.09.002. Epub 2013 Sep 25. Mutat Res Rev Mutat Res. 2014. PMID: 24076409 Review.
Cited by
-
Difluoromethylornithine (DFMO) and AMXT 1501 inhibit capsule biosynthesis in pneumococci.Sci Rep. 2022 Jul 12;12(1):11804. doi: 10.1038/s41598-022-16007-7. Sci Rep. 2022. PMID: 35821246 Free PMC article.
-
A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies.Molecules. 2020 Mar 3;25(5):1135. doi: 10.3390/molecules25051135. Molecules. 2020. PMID: 32138329 Free PMC article. Review.
-
A new 68Ga-labeled ornithine derivative for PET imaging of ornithine metabolism in tumors.Amino Acids. 2023 May;55(5):595-606. doi: 10.1007/s00726-023-03250-z. Epub 2023 Feb 21. Amino Acids. 2023. PMID: 36809562
-
Roles of the components of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori.Future Microbiol. 2024;19(14):1253-1267. doi: 10.1080/17460913.2024.2383514. Epub 2024 Aug 22. Future Microbiol. 2024. PMID: 39171625 Free PMC article. Review.
-
Preventing Gastric Cancer Development by Inhibiting the Virulence of H. pylori Infection.Oncology (Williston Park). 2019 Jun 19;33(6):227-31. Oncology (Williston Park). 2019. PMID: 31219607 Free PMC article.
References
-
- Hooi JKY, et al. Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. - PubMed
-
- Parsonnet J, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. - PubMed
-
- Uemura N, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

