Polo-like kinase 4 maintains centriolar satellite integrity by phosphorylation of centrosomal protein 131 (CEP131)
- PMID: 30804208
- PMCID: PMC6484138
- DOI: 10.1074/jbc.RA118.004867
Polo-like kinase 4 maintains centriolar satellite integrity by phosphorylation of centrosomal protein 131 (CEP131)
Abstract
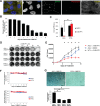
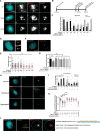
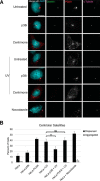
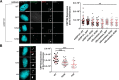
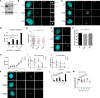
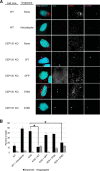
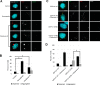
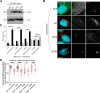
The centrosome, consisting of two centrioles surrounded by a dense network of proteins, is the microtubule-organizing center of animal cells. Polo-like kinase 4 (PLK4) is a Ser/Thr protein kinase and the master regulator of centriole duplication, but it may play additional roles in centrosome function. To identify additional proteins regulated by PLK4, we generated an RPE-1 human cell line with a genetically engineered "analog-sensitive" PLK4AS, which genetically encodes chemical sensitivity to competitive inhibition via a bulky ATP analog. We used this transgenic line in an unbiased multiplex phosphoproteomic screen. Several hits were identified and validated as direct PLK4 substrates by in vitro kinase assays. Among them, we confirmed Ser-78 in centrosomal protein 131 (CEP131, also known as AZI1) as a direct substrate of PLK4. Using immunofluorescence microscopy, we observed that although PLK4-mediated phosphorylation of Ser-78 is dispensable for CEP131 localization, ciliogenesis, and centriole duplication, it is essential for maintaining the integrity of centriolar satellites. We also found that PLK4 inhibition or use of a nonphosphorylatable CEP131 variant results in dispersed centriolar satellites. Moreover, replacement of endogenous WT CEP131 with an S78D phosphomimetic variant promoted aggregation of centriolar satellites. We conclude that PLK4 phosphorylates CEP131 at Ser-78 to maintain centriolar satellite integrity.
Keywords: CEP131; PLK4; cell division; centriolar satellite; centriole; centriole duplication; centrosome; chemical biology; chemical genetics; chromosome segregation; mass spectrometry (MS); microtubule; phosphoproteomics.
© 2019 Denu et al.
Conflict of interest statement
M. E. B. is on the medical advisory board of Strata Oncology and receives research funding from Abbvie, Genentech, Puma, and Loxo Oncology
Figures

Similar articles
-
A non-canonical function of Plk4 in centriolar satellite integrity and ciliogenesis through PCM1 phosphorylation.EMBO Rep. 2016 Mar;17(3):326-37. doi: 10.15252/embr.201541432. Epub 2016 Jan 11. EMBO Rep. 2016. PMID: 26755742 Free PMC article.
-
The centriolar satellite protein Cep131 is important for genome stability.J Cell Sci. 2012 Oct 15;125(Pt 20):4770-9. doi: 10.1242/jcs.104059. Epub 2012 Jul 13. J Cell Sci. 2012. PMID: 22797915
-
The Unkempt RNA-binding protein reveals a local translation program in centriole overduplication.J Cell Biol. 2025 Aug 4;224(8):e202407196. doi: 10.1083/jcb.202407196. Epub 2025 Jul 23. J Cell Biol. 2025. PMID: 40699149
-
Role of Polo-like Kinases Plk1 and Plk4 in the Initiation of Centriole Duplication-Impact on Cancer.Cells. 2022 Feb 24;11(5):786. doi: 10.3390/cells11050786. Cells. 2022. PMID: 35269408 Free PMC article. Review.
-
The PLK4-STIL-SAS-6 module at the core of centriole duplication.Biochem Soc Trans. 2016 Oct 15;44(5):1253-1263. doi: 10.1042/BST20160116. Biochem Soc Trans. 2016. PMID: 27911707 Free PMC article. Review.
Cited by
-
The centrosomal protein 131 participates in the regulation of mitochondrial apoptosis.Commun Biol. 2023 Dec 15;6(1):1271. doi: 10.1038/s42003-023-05676-3. Commun Biol. 2023. PMID: 38102401 Free PMC article.
-
The SON RNA splicing factor is required for intracellular trafficking structures that promote centriole assembly and ciliogenesis.Mol Biol Cell. 2021 Oct 1;32(20):ar4. doi: 10.1091/mbc.E21-06-0305. Epub 2021 Aug 18. Mol Biol Cell. 2021. PMID: 34406792 Free PMC article.
-
Doryphagy: when selective autophagy safeguards centrosome integrity.Mol Cell Oncol. 2020 Feb 7;7(2):1719021. doi: 10.1080/23723556.2020.1719021. eCollection 2020. Mol Cell Oncol. 2020. PMID: 32158930 Free PMC article.
-
CEP131 Abrogates CHK1 Inhibitor-Induced Replication Defects and Is Associated with Unfavorable Outcome in Neuroblastoma.J Oncol. 2020 Sep 15;2020:2752417. doi: 10.1155/2020/2752417. eCollection 2020. J Oncol. 2020. PMID: 33014050 Free PMC article.
-
Proteomic Signature of Host Response to SARS-CoV-2 Infection in the Nasopharynx.Mol Cell Proteomics. 2021;20:100134. doi: 10.1016/j.mcpro.2021.100134. Epub 2021 Aug 14. Mol Cell Proteomics. 2021. PMID: 34400346 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

